Encoder-decoder convolutional neural network for simple CT segmentation of COVID-19 infected lungs
- PMID: 39145207
- PMCID: PMC11323195
- DOI: 10.7717/peerj-cs.2178
Encoder-decoder convolutional neural network for simple CT segmentation of COVID-19 infected lungs
Abstract
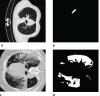
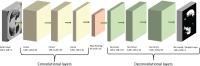
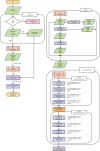
This work presents the application of an Encoder-Decoder convolutional neural network (ED-CNN) model to automatically segment COVID-19 computerised tomography (CT) data. By doing so we are producing an alternative model to current literature, which is easy to follow and reproduce, making it more accessible for real-world applications as little training would be required to use this. Our simple approach achieves results comparable to those of previously published studies, which use more complex deep-learning networks. We demonstrate a high-quality automated segmentation prediction of thoracic CT scans that correctly delineates the infected regions of the lungs. This segmentation automation can be used as a tool to speed up the contouring process, either to check manual contouring in place of a peer checking, when not possible or to give a rapid indication of infection to be referred for further treatment, thus saving time and resources. In contrast, manual contouring is a time-consuming process in which a professional would contour each patient one by one to be later checked by another professional. The proposed model uses approximately 49 k parameters while others average over 1,000 times more parameters. As our approach relies on a very compact model, shorter training times are observed, which make it possible to easily retrain the model using other data and potentially afford "personalised medicine" workflows. The model achieves similarity scores of Specificity (Sp) = 0.996 ± 0.001, Accuracy (Acc) = 0.994 ± 0.002 and Mean absolute error (MAE) = 0.0075 ± 0.0005.
Keywords: Autoencoder; Automated segmentation; CNN; COVID-19; Encoder-decoder; Lung CT; Lung segmentation; Machine learning; Simple segmentation.
© 2024 Newson et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Cheng MM, Fan DP. Structure-measure: a new way to evaluate foreground maps. International Journal of Computer Vision. 2021;129(9):2622–2638. doi: 10.1007/s11263-021-01490-8. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
