Population-level task-evoked functional connectivity via Fourier analysis
- PMID: 39145309
- PMCID: PMC11321825
- DOI: 10.1093/jrsssc/qlae015
Population-level task-evoked functional connectivity via Fourier analysis
Abstract
Functional magnetic resonance imaging (fMRI) is a noninvasive and in-vivo imaging technique essential for measuring brain activity. Functional connectivity is used to study associations between brain regions, either while study subjects perform tasks or during periods of rest. In this paper, we propose a rigorous definition of task-evoked functional connectivity at the population level (ptFC). Importantly, our proposed ptFC is interpretable in the context of task-fMRI studies. An algorithm for estimating the ptFC is provided. We present the performance of the proposed algorithm compared to existing functional connectivity frameworks using simulations. Lastly, we apply the proposed algorithm to estimate the ptFC in a motor-task study from the Human Connectome Project.
Keywords: AMUSE algorithm; Human Connectome Project; motor-task; weakly stationary with mean zero.
© The Royal Statistical Society 2024. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest: None declared.
Figures

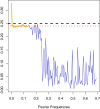
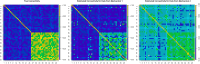

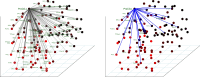
References
-
- Ashby F. G. (2019). Statistical analysis of fMRI data. MIT Press.
-
- Barch D. M., Burgess G. C., Harms M. P., Petersen S. E., Schlaggar B. L., Corbetta M., Glasser M. F., Curtiss S., Dixit S., Feldt C., & Nolan D. (2013). Function in the human connectome: Task-fMRI and individual differences in behavior. Neuroimage, 80, 169–189. 10.1016/j.neuroimage.2013.05.033 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
