Mendelian randomization of plasma lipidome, inflammatory proteome and heart failure
- PMID: 39145416
- PMCID: PMC11631237
- DOI: 10.1002/ehf2.14997
Mendelian randomization of plasma lipidome, inflammatory proteome and heart failure
Abstract
Aims: Heart failure (HF) is a global health issue, with lipid metabolism and inflammation critically implicated in its progression. This study harnesses cutting-edge, expanded genetic information for lipid and inflammatory protein profiles, employing Mendelian randomization (MR) to uncover genetic risk factors for HF.
Methods: We assessed genetic susceptibility to HF across 179 lipidomes and 91 inflammatory proteins using instrumental variables (IVs) from recent genome-wide association studies (GWASs) and proteome-wide quantitative trait loci (pQTL) studies. GWASs involving 47 309 HF cases and 930 014 controls were obtained from the Heart Failure Molecular Epidemiology for Therapeutic Targets (HERMES) Consortium. Data on 179 lipids from 7174 individuals in a Finnish cohort and 91 inflammatory proteins from a European pQTL study involving 14 824 individuals are available in the HGRI-EBI catalogue. A two-sample MR approach evaluated the associations, and a two-step mediation analysis explored the mediation role of inflammatory proteins in the lipid-HF pathway. Sensitivity analyses, including MR-RAPS (robust adjusted profile score) and MR-Egger, ensured result robustness.
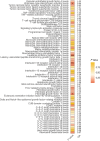
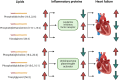
Results: Genetic IVs for 162 lipids and 74 inflammatory proteins were successfully identified. MR analysis revealed a genetic association between HF and 31 lipids. Among them, 18 lipids, including sterol ester (27:1/18:0), cholesterol, 9 phosphatidylcholines, phosphatidylinositol (16:0_20:4) and 6 triacylglycerols, were identified as HF risk factors [odds ratio (OR) = 1.037-1.368]. Cholesterol exhibited the most significant association with elevated HF risk [OR = 1.368, 95% confidence interval (CI) = 1.044-1.794, P = 0.023]. In the inflammatory proteome, leukaemia inhibitory factor receptor (OR = 0.841, 95% CI = 0.789-0.897, P = 1.08E-07), fibroblast growth factor 19 (OR = 0.905, 95% CI = 0.830-0.988, P = 0.025) and urokinase-type plasminogen activator (OR = 0.938, 95% CI = 0.886-0.994, P = 0.030) were causally negatively correlated with HF, whereas interleukin-20 receptor subunit alpha (OR = 1.333, 95% CI = 1.094-1.625, P = 0.004) was causally positively correlated with HF. Mediation analysis revealed leukaemia inhibitory factor receptor (mediation proportion: 23.5%-25.2%) and urokinase-type plasminogen activator (mediation proportion: 9.5%-10.7%) as intermediaries in the lipid-inflammation-HF pathway. No evidence of directional horizontal pleiotropy was observed (P > 0.05).
Conclusions: This study identifies a genetic connection between certain lipids, particularly cholesterol, and HF, highlighting inflammatory proteins that influence HF risk and mediate this relationship, suggesting new therapeutic targets and insights into genetic drivers in HF.
Keywords: Mendelian randomization; causality; heart failure; inflammatory proteome; lipidome; mediator.
© 2024 The Author(s). ESC Heart Failure published by John Wiley & Sons Ltd on behalf of European Society of Cardiology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Causal correlations between inflammatory proteins and heart failure: A two-sample Mendelian randomization analysis.ESC Heart Fail. 2025 Apr;12(2):1374-1385. doi: 10.1002/ehf2.15151. Epub 2024 Nov 6. ESC Heart Fail. 2025. PMID: 39501838 Free PMC article.
-
Genetic causal association between lipidomic profiles, inflammatory proteomics, and aortic stenosis: a Mendelian randomization investigation.Eur J Med Res. 2024 Aug 31;29(1):446. doi: 10.1186/s40001-024-02014-z. Eur J Med Res. 2024. PMID: 39217396 Free PMC article.
-
Mendelian randomization of circulating proteome identifies actionable targets in heart failure.BMC Genomics. 2022 Aug 13;23(1):588. doi: 10.1186/s12864-022-08811-2. BMC Genomics. 2022. PMID: 35964012 Free PMC article.
-
Proteome-wide mendelian randomization investigates potential associations in heart failure and its etiology: emphasis on PCSK9.BMC Med Genomics. 2024 Feb 21;17(1):59. doi: 10.1186/s12920-024-01826-6. BMC Med Genomics. 2024. PMID: 38383373 Free PMC article.
-
Evaluating the association between lipidome and female reproductive diseases through comprehensive Mendelian randomization analyses.Sci Rep. 2025 Jan 19;15(1):2448. doi: 10.1038/s41598-025-86794-2. Sci Rep. 2025. PMID: 39828767 Free PMC article.
Cited by
-
Dissecting causal networks of inflammatory factors and metabolites in heart failure: A mediation Mendelian randomization study.Medicine (Baltimore). 2025 Aug 8;104(32):e43801. doi: 10.1097/MD.0000000000043801. Medicine (Baltimore). 2025. PMID: 40797443 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

