Discovery of Potent Degraders of the Dengue Virus Envelope Protein
- PMID: 39145423
- PMCID: PMC11516100
- DOI: 10.1002/advs.202405829
Discovery of Potent Degraders of the Dengue Virus Envelope Protein
Abstract
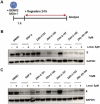
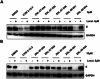
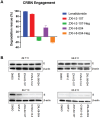
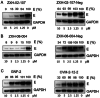
Targeted protein degradation has been widely adopted as a new approach to eliminate both established and previously recalcitrant therapeutic targets. Here, it is reported that the development of small molecule degraders of the envelope (E) protein of dengue virus. Two classes of bivalent E-degraders are developed by linking two previously reported E-binding small molecules, GNF-2, and CVM-2-12-2, to a glutarimide-based recruiter of the CRL4CRBN ligase to effect proteosome-mediated degradation of the E protein. ZXH-2-107 (based on GNF-2) is an E-degrader with ABL inhibitory activity while ZXH-8-004 (based on CVM-2-12-2) is a selective and potent E-degrader. These two compounds provide proof of concept that difficult-to-drug targets such as a viral envelope protein can be effectively eliminated using a bivalent degrader and provide starting points for the future development of a new class of direct-acting antiviral drugs.
Keywords: antivirals; dengue; envelope protein; infection; protein degradation.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
N.S.G. is a Scientific Founder, member of the SAB, and equity holder in C4 Therapeutics, Syros, Soltego (board member), Voronoi, Allorion, Lighthorse, GSK, Larkspur (board member), Shenandoah (board member) and Matchpoint. The Gray lab receives research funding from Springworks and Simcere. T.Z. is a scientific founder, equity holder, and consultant of Matchpoint, equity holder of Shenandoah. J.C. is a co‐founder and equality holder of Matchpoint Therapeutics, a scientific co‐founder of M3 Bioinformatics & Technology Inc., and a consultant and equity holder for Soltego and Allorion. E.S.F. is a founder, member of the scientific advisory board (SAB), and equity holder of Civetta Therapeutics, Lighthorse, Proximity Therapeutics, and Neomorph Inc (also board of directors), SAB member and equity holder in Avilar Therapeutics and Photys Therapeutics, and a consultant to Astellas, Sanofi, Novartis, Deerfield, Ajax and EcoR1 capital. The Fischer laboratory receives or has received research funding from Novartis, Deerfield, Ajax, Interline, and Astellas. K.A.D receives or has received consulting fees from Kronos Bio and Neomorph Inc.
Figures

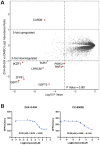
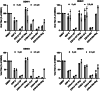
Update of
-
Discovery of Potent Degraders of the Dengue Virus Envelope Protein.bioRxiv [Preprint]. 2024 Jun 2:2024.06.01.596987. doi: 10.1101/2024.06.01.596987. bioRxiv. 2024. Update in: Adv Sci (Weinh). 2024 Oct;11(40):e2405829. doi: 10.1002/advs.202405829. PMID: 38854003 Free PMC article. Updated. Preprint.
References
-
- a) Simmonds P., Becher P., Bukh J., Gould E. A., Meyers G., Monath T., Muerhoff S., Pletnev A., Rico‐Hesse R., Smith D. B., Stapleton J. T., Ictv Report C., J. Gen. Virol. 2017, 98, 2; - PMC - PubMed
- b) Ehrenkranz N. J., Ventura A. K., Cuadrado R. R., Pond W. L., Porter J. E., N. Engl. J. Med. 1971, 285, 1460. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
