XOL-1 regulates developmental timing by modulating the H3K9 landscape in C. elegans early embryos
- PMID: 39146391
- PMCID: PMC11349215
- DOI: 10.1371/journal.pgen.1011238
XOL-1 regulates developmental timing by modulating the H3K9 landscape in C. elegans early embryos
Abstract
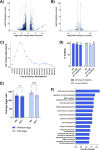
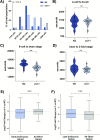
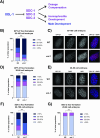
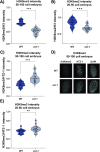
Sex determination in the nematode C. elegans is controlled by the master regulator XOL-1 during embryogenesis. Expression of xol-1 is dependent on the ratio of X chromosomes and autosomes, which differs between XX hermaphrodites and XO males. In males, xol-1 is highly expressed and in hermaphrodites, xol-1 is expressed at very low levels. XOL-1 activity is known to be critical for the proper development of C. elegans males, but its low expression was considered to be of minimal importance in the development of hermaphrodite embryos. Our study reveals that XOL-1 plays an important role as a regulator of developmental timing during hermaphrodite embryogenesis. Using a combination of imaging and bioinformatics techniques, we found that hermaphrodite embryos have an accelerated rate of cell division, as well as a more developmentally advanced transcriptional program when xol-1 is lost. Further analyses reveal that XOL-1 is responsible for regulating the timing of initiation of dosage compensation on the X chromosomes, and the appropriate expression of sex-biased transcriptional programs in hermaphrodites. We found that xol-1 mutant embryos overexpress the H3K9 methyltransferase MET-2 and have an altered H3K9me landscape. Some of these effects of the loss of xol-1 gene were reversed by the loss of met-2. These findings demonstrate that XOL-1 plays an important role as a developmental regulator in embryos of both sexes, and that MET-2 acts as a downstream effector of XOL-1 activity in hermaphrodites.
Copyright: © 2024 Jash et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Wolff JR, Zarkower D. Chapter 1 Somatic Sexual Differentiation in Caenorhabditis elegans. In: Sex Determination and Sexual Development [Internet]. Academic Press; 2008. p. 1–39. (Current Topics in Developmental Biology; vol. 83). Available from: https://www.sciencedirect.com/science/article/pii/S0070215308004018. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

