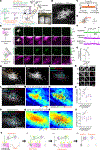
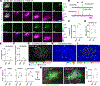
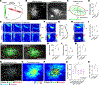
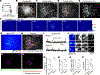
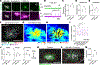
Fig. 1.. Synchronized firing between individual RGC axons and retinal waves instructs axon branch dynamics.
(A) Schematic of the in vivo two-photon (2P) imaging approach for tracking axon branch dynamics of a single RGC and simultaneous dual-color calcium imaging of the RGC axon activity and retinal waves in the SC of awake, head-restrained mice at P8 to P9. Stochastic expression of Cre in a few RGCs was achieved by intravitreal injections of AAV2/1-TRE-Cre in Ai162 mice, which harbor CAG-LSL-tTA2 and TRE2-LSL-GCaMP6s alleles. AAV2/2-CAG-FLEX-EGFP and AAV2/1-Syn-jRGECO1a were also injected intravitreally. As a result, Cre+ RGCs expressed GFP, GCaMP6s, and jRGECO1a, whereas Cre− RGCs only expressed jRGECO1a. The blue-boxed area is magnified in (B). Scale bar, 500 μm. PMT, photomultiplier tube; Ti:Sapphire, Ti:Sapphire laser. (B) In vivo two-photon imaging of an entire single RGC axon within a depth of 200 μm below the surface of the SC at P8. Directions R, L, M, and C correspond to rostral, lateral, medial, and caudal in the SC, respectively, unless otherwise stated. Scale bar, 100 μm. (C) Experimental timeline. Z stacks of a single–RGC axon arbor were acquired at a 2-hour interval, and dual-color calcium imaging was performed for 90 min in between taking the z stacks. (D) Traced z projection of axon branches from a single RGC within ±20 μm of the optical plane for dual-color calcium imaging. Orange and red circles indicate central and distal regions of interest (ROIs) for calculating the retinal wave traces shown in (F), respectively. (E) Example ΔF/F (fractional change in fluorescence) montages of single-axon firing (GCaMP6s) and of retinal waves (jRGECO1a). In montage 1, the axon did not fire when a retinal wave partially overlapped with its axon arbors. In montage 2, the axon fired when a retinal wave passed the center of the axon arbor. Orange and red circles indicate ROIs in the central and distal regions of the axon, respectively. Scale bar, 100 μm. (F) A GCaMP6s signal trace (ΔF/F) from the single RGC axon and jRGECO1a signal traces (ΔF/F) from the ROIs indicated in (D). The periods (1 and 2) depicted in montages (E) are shown in gray. (G) The fraction of synchronization between single-axon firing and retinal waves at central regions of the axons was significantly higher than that at distal regions. Data points from the same axon were paired (central, 0.92 ± 0.02; distal, 0.49 ± 0.05; **P = 0.008, one-tailed Wilcoxon signed-rank test, n = 7 axons from 7 animals). (H and I) Z projection of a single RGC axon at 0 (H) and 2 hours (I) of in vivo two-photon imaging. The orange-boxed areas in (H) are magnified in (K). Scale bar, 100 μm. (J) A single reconstructed RGC axon. The behavior of individual branches over the 2-hour interval were categorized as added (green), eliminated (red), extended (cyan), and retracted (magenta). (K) Zoomed-in images [corresponding to numbered orange boxes in (H)] show changes of axon branches over the 2-hour imaging session. The left and middle columns show projected images of two to three optical sections with 2-μm intervals at 0 and 2 hours. The right column displays changes of branch dynamics in 2 hours. Scale bar, 10 μm. (L) Terminals of stable (white), eliminated (red), extended (cyan), and retracted (magenta) axon branches at 2 hours were labeled on a z projection of the single–RGC axon arbor. The positions from which newly added branches emerged were also labeled (green). Scale bar, 100 μm. (M) A spatial map of Pearson’s correlation coefficients (r) between the activity of the single RGC axon and retinal waves. (N) Stable and eliminated branch terminals and positions where newly added branches emerged from were plotted on the correlation map. (O) Means of correlation coefficients [stable, 0.34 ± 0.01; added, 0.42 ± 0.01, P = 0.0004; eliminated, 0.26 ± 0.03, P = 0.001; extended, 0.33 ± 0.02, P = 0.999; retracted, 0.31 ± 0.03, P = 0.36; ns, not significant; one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison test, n = 7 axons from 7 animals]. (P to S) Simultaneous in vivo two-photon imaging of single–RGC axon branch dynamics and dual-color calcium imaging of axonal activity and postsynaptic waves in the SC. Stochastic expression of Cre in a few RGCs was achieved by intravitreal injections of AAV2/1-TRE-Cre in Ai162 mice, which harbor CAG-LSL-tTA2 and TRE-LSL-GCaMP6s alleles, and expression of jRGECO1a in neurons of the SC was achieved by injection of AAV2/9-Syn-NES-jRGECO1a into the SC one day after eye injection. After the AAV injections, only a few RGC axons expressed GFP and GCaMP6, and most of SC neurons expressed jRGECO1a. (P) Terminals of stable, eliminated, extended, and retracted axon branches at 2 hours were labeled on a z projection of the single RGC axon arbor. The positions from which newly added branches emerged were also labeled. Scale bar, 100 mm. (Q) A spatial map of Pearson’s correlation coefficients between presynaptic activity of the single–RGC axon arbor and postsynaptic waves. (R) Stable and eliminated branch terminals and positions from where newly added branches emerged were plotted on the correlation map. (S) Mean correlation coefficients (stable, 0.52 ± 0.03; added, 0.57 ± 0.03, P = 0.0097; eliminated, 0.44 ± 0.03, P = 0.0002; extended, 0.49 ± 0.03, P = 0.29; retracted, 0.51 ± 0.03, P = 0.88, one-way ANOVA with Dunnett’s multiple comparison test, n = 6 axons from 6 animals). (T) Schematic model of Hebbian axon remodeling by endogenous patterns of spontaneous activity. Synchronized presynaptic inputs (retinal waves) excite postsynaptic cells (i and ii). When an RGC axon takes part in firing a SC neuron repeatedly or persistently (i), the axon forms new branches near the SC neurons to increase their efficacy (iii). By contrast, when an RGC axon does not participate in making an SC neuron fire repeatedly or persistently (ii), branches of the axon near the SC neuron are eliminated (iii). For all the box plots, the central line indicates the median, and the bottom and top edges indicate the 25th and 75th percentiles of the data across animals, respectively. **P < 0.01; ***P < 0.001; ns, not significant. Data are mean ± SEM.