Maternal antibiotic prophylaxis during cesarean section has a limited impact on the infant gut microbiome
- PMID: 39146801
- PMCID: PMC11335186
- DOI: 10.1016/j.chom.2024.07.010
Maternal antibiotic prophylaxis during cesarean section has a limited impact on the infant gut microbiome
Abstract
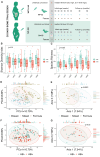
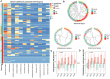
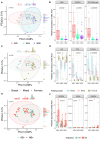
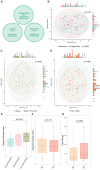
Pregnant women undergoing a cesarean section (CS) typically receive antibiotics prior to skin incision to prevent infections. To investigate if the timing of antibiotics influences the infant gut microbiome, we conducted a randomized controlled trial (NCT06030713) in women delivering via a scheduled CS who received antibiotics either before skin incision or after umbilical cord clamping. We performed a longitudinal analysis on 172 samples from 28 infants at 8 post-birth time points and a cross-sectional analysis at 1 month in 79 infants from 3 cohorts. Although no significant associations with bacterial composition, metabolic pathways, short-chain fatty acids, and bile acids were found, we observed subtle differences between the groups at the bacterial strain level and in the load of antibiotic resistance genes. Rather, feeding mode was a predominant and defining factor impacting infant microbial composition. In conclusion, antibiotic administration during CS has only limited effects on the early-life gut microbiome.
Keywords: antibiotic resistance genes; cesarean section; feeding mode; infant gut microbiome; maternal prophylactic antibiotics; resistome; strain variability.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.d.M. has served as a speaker for Danone Nutricia Research and Mead Johnson. The funders had no role in study design, data analysis, data interpretation, writing of the manuscript, and the decision to publish.
Figures

References
-
- Yassour M., Vatanen T., Siljander H., Hämäläinen A.-M., Härkönen T., Ryhänen S.J., Franzosa E.A., Vlamakis H., Huttenhower C., Gevers D., et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aad0917. - DOI - PMC - PubMed
-
- National Institute for Health and Clinical Excellence . 2021. NICE guideline [NG192]https://www.nice.org.uk/guidance/ng192 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

