Structure of a fully assembled γδ T cell antigen receptor
- PMID: 39146975
- PMCID: PMC11485255
- DOI: 10.1038/s41586-024-07920-0
Structure of a fully assembled γδ T cell antigen receptor
Abstract
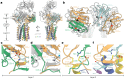
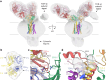
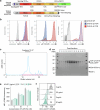
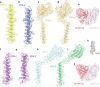
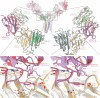
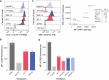
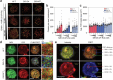
T cells in jawed vertebrates comprise two lineages, αβ T cells and γδ T cells, defined by the antigen receptors they express-that is, αβ and γδ T cell receptors (TCRs), respectively. The two lineages have different immunological roles, requiring that γδ TCRs recognize more structurally diverse ligands1. Nevertheless, the receptors use shared CD3 subunits to initiate signalling. Whereas the structural organization of αβ TCRs is understood2,3, the architecture of γδ TCRs is unknown. Here, we used cryogenic electron microscopy to determine the structure of a fully assembled, MR1-reactive, human Vγ8Vδ3 TCR-CD3δγε2ζ2 complex bound by anti-CD3ε antibody Fab fragments4,5. The arrangement of CD3 subunits in γδ and αβ TCRs is conserved and, although the transmembrane α-helices of the TCR-γδ and -αβ subunits differ markedly in sequence, packing of the eight transmembrane-helix bundles is similar. However, in contrast to the apparently rigid αβ TCR2,3,6, the γδ TCR exhibits considerable conformational heterogeneity owing to the ligand-binding TCR-γδ subunits being tethered to the CD3 subunits by their transmembrane regions only. Reducing this conformational heterogeneity by transfer of the Vγ8Vδ3 TCR variable domains to an αβ TCR enhanced receptor signalling, suggesting that γδ TCR organization reflects a compromise between efficient signalling and the ability to engage structurally diverse ligands. Our findings reveal the marked structural plasticity of the TCR on evolutionary timescales, and recast it as a highly versatile receptor capable of initiating signalling as either a rigid or flexible structure.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

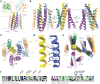








References
-
- Willcox, B. E. & Willcox, C. R. γδ TCR ligands: the quest to solve a 500-million-year-old mystery. Nat. Immunol.20, 121–128 (2019). - PubMed
-
- Dong, D. et al. Structural basis of assembly of the human T cell receptor-CD3 complex. Nature573, 546–552 (2019). - PubMed
-
- Beverley, P. C. & Callard, R. E. Distinctive functional characteristics of human “T” lymphocytes defined by E rosetting or a monoclonal anti-T cell antibody. Eur. J. Immunol.11, 329–334 (1981). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

