Transporter-mediated depletion of extracellular proline directly contributes to plant pattern-triggered immunity against a bacterial pathogen
- PMID: 39147739
- PMCID: PMC11327374
- DOI: 10.1038/s41467-024-51244-6
Transporter-mediated depletion of extracellular proline directly contributes to plant pattern-triggered immunity against a bacterial pathogen
Abstract
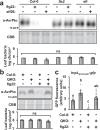
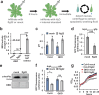
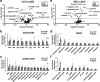
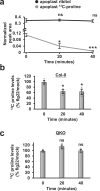
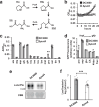
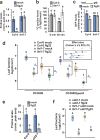
Plants possess cell surface-localized immune receptors that detect microbe-associated molecular patterns (MAMPs) and initiate defenses that provide effective resistance against microbial pathogens. Many MAMP-induced signaling pathways and cellular responses are known, yet how pattern-triggered immunity (PTI) limits pathogen growth in plants is poorly understood. Through a combined metabolomics and genetics approach, we discovered that plant-exuded proline is a virulence-inducing signal and nutrient for the bacterial pathogen Pseudomonas syringae, and that MAMP-induced depletion of proline from the extracellular spaces of Arabidopsis leaves directly contributes to PTI against P. syringae. We further show that MAMP-induced depletion of extracellular proline requires the amino acid transporter Lysine Histidine Transporter 1 (LHT1). This study demonstrates that depletion of a single extracellular metabolite is an effective component of plant induced immunity. Given the important role for amino acids as nutrients for microbial growth, their depletion at sites of infection may be a broadly effective means for defense against many pathogens.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

