FIGNL1-FIRRM is essential for meiotic recombination and prevents DNA damage-independent RAD51 and DMC1 loading
- PMID: 39147779
- PMCID: PMC11327267
- DOI: 10.1038/s41467-024-51458-8
FIGNL1-FIRRM is essential for meiotic recombination and prevents DNA damage-independent RAD51 and DMC1 loading
Abstract
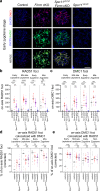
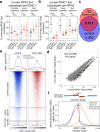
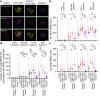
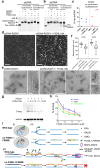
During meiosis, nucleoprotein filaments of the strand exchange proteins RAD51 and DMC1 are crucial for repairing SPO11-generated DNA double-strand breaks (DSBs) by homologous recombination (HR). A balanced activity of positive and negative RAD51/DMC1 regulators ensures proper recombination. Fidgetin-like 1 (FIGNL1) was previously shown to negatively regulate RAD51 in human cells. However, FIGNL1's role during meiotic recombination in mammals remains unknown. Here, we decipher the meiotic functions of FIGNL1 and FIGNL1 Interacting Regulator of Recombination and Mitosis (FIRRM) using male germline-specific conditional knock-out (cKO) mouse models. Both FIGNL1 and FIRRM are required for completing meiotic prophase in mouse spermatocytes. Despite efficient recruitment of DMC1 on ssDNA at meiotic DSB hotspots, the formation of late recombination intermediates is defective in Firrm cKO and Fignl1 cKO spermatocytes. Moreover, the FIGNL1-FIRRM complex limits RAD51 and DMC1 accumulation on intact chromatin, independently from the formation of SPO11-catalyzed DSBs. Purified human FIGNL1ΔN alters the RAD51/DMC1 nucleoprotein filament structure and inhibits strand invasion in vitro. Thus, this complex might regulate RAD51 and DMC1 association at sites of meiotic DSBs to promote proficient strand invasion and processing of recombination intermediates.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
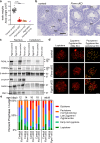
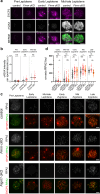
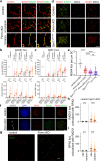
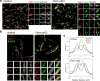
References
MeSH terms
Substances
Grants and funding
- ANR-17-426 CE12-0015/Agence Nationale de la Recherche (French National Research Agency)
- ANR-17-426 CE12-0015/Agence Nationale de la Recherche (French National Research Agency)
- ANR-17-426 CE12-0015/Agence Nationale de la Recherche (French National Research Agency)
- ANR-17-426 CE12-0015/Agence Nationale de la Recherche (French National Research Agency)
- ANR-17-426 CE12-0015/Agence Nationale de la Recherche (French National Research Agency)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

