LIFR regulates cholesterol-driven bidirectional hepatocyte-neutrophil cross-talk to promote liver regeneration
- PMID: 39147934
- PMCID: PMC11498095
- DOI: 10.1038/s42255-024-01110-y
LIFR regulates cholesterol-driven bidirectional hepatocyte-neutrophil cross-talk to promote liver regeneration
Abstract
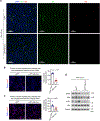
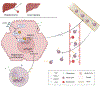
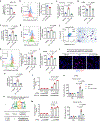
Liver regeneration is under metabolic and immune regulation. Despite increasing recognition of the involvement of neutrophils in regeneration, it is unclear how the liver signals to the bone marrow to release neutrophils after injury and how reparative neutrophils signal to hepatocytes to reenter the cell cycle. Here we report that loss of the liver tumour suppressor Lifr in mouse hepatocytes impairs, whereas overexpression of leukaemia inhibitory factor receptor (LIFR) promotes liver repair and regeneration after partial hepatectomy or toxic injury. In response to physical or chemical damage to the liver, LIFR from hepatocytes promotes the secretion of cholesterol and CXCL1 in a STAT3-dependent manner, leading to the efflux of bone marrow neutrophils to the circulation and damaged liver. Cholesterol, via its receptor ERRα, stimulates neutrophils to secrete hepatocyte growth factor to accelerate hepatocyte proliferation. Altogether, our findings reveal a LIFR-STAT3-CXCL1-CXCR2 axis and a LIFR-STAT3-cholesterol-ERRα-hepatocyte growth factor axis that form bidirectional hepatocyte-neutrophil cross-talk to repair and regenerate the liver.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
H.Z. consults for Flagship Pioneering, Alnylam Pharmaceuticals, Jumble Therapeutics, and Chroma Medicines, and serves on the SAB of Ubiquitix. H.Z. has research support from Chroma Medicines. H.Z. owns stock in Ionis and Madrigal Pharmaceuticals. M.A.C. reports grants, personal fees, and an ownership interest in ImmunoGenesis, Inc. and personal fees from AstraZeneca, Inc. The above interests are not directly related to the contents of this paper. The other authors declare no competing interests.
Figures















References
-
- Michalopoulos GK & Bhushan B Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol 18, 40–55 (2021). - PubMed
-
- Forbes SJ & Rosenthal N Preparing the ground for tissue regeneration: from mechanism to therapy. Nat Med 20, 857–869 (2014). - PubMed
-
- Sakamoto Y et al. Favorable indications for hepatectomy in patients with liver metastasis from gastric cancer. J Surg Oncol 95, 534–539 (2007). - PubMed
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- R01AA028791/U.S. Department of Health & Human Services | NIH | National Institute on Alcohol Abuse and Alcoholism (NIAAA)
- DBG-22-161-01-MM/American Cancer Society (American Cancer Society, Inc.)
- R01CA269140/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01DK125396/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01 AA028791/AA/NIAAA NIH HHS/United States
- R01CA166051/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 DK125396/DK/NIDDK NIH HHS/United States
- R01 CA166051/CA/NCI NIH HHS/United States
- RP220614/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)
- R01 CA269140/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

