This is a preprint.
Loss-of-function of the Zinc Finger Homeobox 4 (ZFHX4) gene underlies a neurodevelopmental disorder
- PMID: 39148819
- PMCID: PMC11326360
- DOI: 10.1101/2024.08.07.24311381
Loss-of-function of the Zinc Finger Homeobox 4 (ZFHX4) gene underlies a neurodevelopmental disorder
Update in
-
Loss of function of the zinc finger homeobox 4 gene, ZFHX4, underlies a neurodevelopmental disorder.Am J Hum Genet. 2025 Jun 5;112(6):1388-1414. doi: 10.1016/j.ajhg.2025.04.008. Epub 2025 May 13. Am J Hum Genet. 2025. PMID: 40367947
Abstract
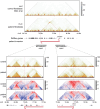
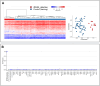
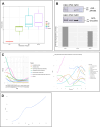
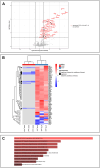
8q21.11 microdeletions encompassing the gene encoding transcription factor ZFHX4, have previously been associated by us with a syndromic form of intellectual disability, hypotonia, decreased balance and hearing loss. Here, we report on 57 individuals, 52 probands and 5 affected family members, with protein truncating variants (n=36), (micro)deletions (n=20) or an inversion (n=1) affecting ZFHX4 with variable developmental delay and intellectual disability, distinctive facial characteristics, morphological abnormalities of the central nervous system, behavioral alterations, short stature, hypotonia, and occasionally cleft palate and anterior segment dysgenesis. The phenotypes associated with 8q21.11 microdeletions and ZFHX4 intragenic loss-of-function variants largely overlap, identifying ZFHX4 as the main driver for the microdeletion syndrome, although leukocyte-derived DNA shows a mild common methylation profile for (micro)deletions only. We identify ZFHX4 as a transcription factor that is increasingly expressed during human brain development and neuronal differentiation. Furthermore, ZFHX4 interacting factors identified via IP-MS in neural progenitor cells, suggest an important role for ZFHX4 in cellular and developmental pathways, especially during histone modifications, cytosolic transport and development. Additionally, using CUT&RUN, we observed that ZFHX4 binds with the promoter regions of genes with crucial roles in embryonic, neuron and axon development. Since loss-of-function variants in ZFHX4 are found with consistent dysmorphic facial features, we investigated whether the disruption of zfhx4 causes craniofacial abnormalities in zebrafish. First-generation (F0) zfhx4 crispant zebrafish, (mosaic) mutant for zfhx4 loss-of-function variants, have significantly shorter Meckel's cartilages and smaller ethmoid plates compared to control zebrafish. Furthermore, behavioral assays show a decreased movement frequency in the zfhx4 crispant zebrafish in comparison with control zebrafish larvae. Although further research is needed, our in vivo work suggests a role for zfhx4 in facial skeleton patterning, palatal development and behavior.
Conflict of interest statement
Declaration of interests No declaration of interests is reported in this manuscript.
Figures



References
-
- Palomares M., Delicado A., Mansilla E., De Torres M.L., Vallespín E., Fernandez L., Martinez-Glez V., García-Miñaur S., Nevado J., Simarro F.S., et al. (2011). Characterization of a 8q21.11 microdeletion syndrome associated with intellectual disability and a recognizable phenotype. American Journal of Human Genetics 89, 295–301. 10.1016/j.ajhg.2011.06.012. - DOI - PMC - PubMed
-
- Vulto-van Silfhout A.T., Hehir-Kwa J.Y., van Bon B.W.M., Schuurs-Hoeijmakers J.H.M., Meader S., Hellebrekers C.J.M., Thoonen I.J.M., de Brouwer A.P.M., Brunner H.G., Webber C., et al. (2013). Clinical significance of de novo and inherited copy-number variation. Hum Mutat 34, 1679–1687. 10.1002/humu.22442. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
