This is a preprint.
Dynamic and prognostic proteomic associations with FEV1 decline in chronic obstructive pulmonary disease
- PMID: 39148837
- PMCID: PMC11326337
- DOI: 10.1101/2024.08.07.24311507
Dynamic and prognostic proteomic associations with FEV1 decline in chronic obstructive pulmonary disease
Abstract
Rationale: Identification and validation of circulating biomarkers for lung function decline in COPD remains an unmet need.
Objective: Identify prognostic and dynamic plasma protein biomarkers of COPD progression.
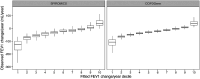
Methods: We measured plasma proteins using SomaScan from two COPD-enriched cohorts, the Subpopulations and Intermediate Outcomes Measures in COPD Study (SPIROMICS) and Genetic Epidemiology of COPD (COPDGene), and one population-based cohort, Multi-Ethnic Study of Atherosclerosis (MESA) Lung. Using SPIROMICS as a discovery cohort, linear mixed models identified baseline proteins that predicted future change in FEV1 (prognostic model) and proteins whose expression changed with change in lung function (dynamic model). Findings were replicated in COPDGene and MESA-Lung. Using the COPD-enriched cohorts, Gene Set Enrichment Analysis (GSEA) identified proteins shared between COPDGene and SPIROMICS. Metascape identified significant associated pathways.
Measurements and main results: The prognostic model found 7 significant proteins in common (p < 0.05) among all 3 cohorts. After applying false discovery rate (adjusted p < 0.2), leptin remained significant in all three cohorts and growth hormone receptor remained significant in the two COPD cohorts. Elevated baseline levels of leptin and growth hormone receptor were associated with slower rate of decline in FEV1. Twelve proteins were nominally but not FDR significant in the dynamic model and all were distinct from the prognostic model. Metascape identified several immune related pathways unique to prognostic and dynamic proteins.
Conclusion: We identified leptin as the most reproducible COPD progression biomarker. The difference between prognostic and dynamic proteins suggests disease activity signatures may be different from prognosis signatures.
Keywords: Disease Progression; Proteomics; Spirometry.
Figures




References
-
- Dransfield MT, Kunisaki KM, Strand MJ, Anzueto A, Bhatt SP, Bowler RP, Criner GJ, Curtis JL, Hanania NA, Nath H, Putcha N, Roark SE, Wan ES, Washko GR, Wells JM, Wendt CH, Make BJ. Acute Exacerbations and Lung Function Loss in Smokers with and without Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med 2017; 195: 324–330. - PMC - PubMed
-
- Kanner RE, Anthonisen NR, Connett JE. Lower respiratory illnesses promote FEV(1) decline in current smokers but not ex-smokers with mild chronic obstructive pulmonary disease: results from the lung health study. Am J Respir Crit Care Med 2001; 164: 358–364. - PubMed
-
- Agusti A, Soriano JB. COPD as a systemic disease. COPD 2008; 5: 133–138. - PubMed
Publication types
Grants and funding
- T32 HL155005/HL/NHLBI NIH HHS/United States
- 75N92020D00001/HL/NHLBI NIH HHS/United States
- N01 HC095167/HL/NHLBI NIH HHS/United States
- HHSN268200900016C/HL/NHLBI NIH HHS/United States
- U01 HL137880/HL/NHLBI NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- HHSN268200900018C/HL/NHLBI NIH HHS/United States
- N01 HC095166/HL/NHLBI NIH HHS/United States
- R01 HL144718/HL/NHLBI NIH HHS/United States
- N01 HC095160/HL/NHLBI NIH HHS/United States
- 75N92020D00002/HL/NHLBI NIH HHS/United States
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- HHSN268200900019C/HL/NHLBI NIH HHS/United States
- U24 HL141762/HL/NHLBI NIH HHS/United States
- N01 HC095161/HL/NHLBI NIH HHS/United States
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- T32 HL144442/HL/NHLBI NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- R01 HL137995/HL/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- P30 ES005605/ES/NIEHS NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- R01 HL077612/HL/NHLBI NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- 75N92020D00003/HL/NHLBI NIH HHS/United States
- R01 HL093081/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- HHSN268200900015C/HL/NHLBI NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- 75N92020D00004/HL/NHLBI NIH HHS/United States
- N01 HC095163/HL/NHLBI NIH HHS/United States
- 75N92020D00007/HL/NHLBI NIH HHS/United States
- R01 HL130506/HL/NHLBI NIH HHS/United States
- HHSN268200900017C/HL/NHLBI NIH HHS/United States
- HHSN268200900020C/HL/NHLBI NIH HHS/United States
- HHSN268200900013C/HL/NHLBI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- 75N92020D00006/HL/NHLBI NIH HHS/United States
- HHSN268200900014C/HL/NHLBI NIH HHS/United States
- N01 HC095165/HL/NHLBI NIH HHS/United States
- N01 HC095164/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
