This is a preprint.
Fast Whole-Brain MR Multi-Parametric Mapping with Scan-Specific Self-Supervised Networks
- PMID: 39148933
- PMCID: PMC11326419
Fast Whole-Brain MR Multi-Parametric Mapping with Scan-Specific Self-Supervised Networks
Abstract
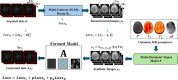
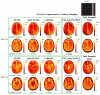
Quantification of tissue parameters using MRI is emerging as a powerful tool in clinical diagnosis and research studies. The need for multiple long scans with different acquisition parameters prohibits quantitative MRI from reaching widespread adoption in routine clinical and research exams. Accelerated parameter mapping techniques leverage parallel imaging, signal modelling and deep learning to offer more practical quantitative MRI acquisitions. However, the achievable acceleration and the quality of maps are often limited. Joint MAPLE is a recent state-of-the-art multi-parametric and scan-specific parameter mapping technique with promising performance at high acceleration rates. It synergistically combines parallel imaging, model-based and machine learning approaches for joint mapping of , proton density and the field inhomogeneity. However, Joint MAPLE suffers from prohibitively long reconstruction time to estimate the maps from a multi-echo, multi-flip angle (MEMFA) dataset at high resolution in a scan-specific manner. In this work, we propose a faster version of Joint MAPLE which retains the mapping performance of the original version. Coil compression, random slice selection, parameter-specific learning rates and transfer learning are synergistically combined in the proposed framework. It speeds-up the reconstruction time up to 700 times than the original version and processes a whole-brain MEMFA dataset in 21 minutes on average, which originally requires ~260 hours for Joint MAPLE. The mapping performance of the proposed framework is ~2-fold better than the standard and the state-of-the-art evaluated reconstruction techniques on average in terms of the root mean squared error.
Keywords: parameter mapping; quantitative MRI; scan-specific deep learning; self-supervised networks.
Figures






References
-
- Arshad M., Qureshi M., Inam O., Omer H., 2021. Transfer learning in deep neural network based under-sampled MR image reconstruction. Magn Reson Imaging 76, 96–107. - PubMed
-
- Block K.T., Uecker M., Frahm J., 2009. Model-based iterative reconstruction for radial fast spin-echo MRI. IEEE Trans Med Imaging 28, 1759–1769. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
