Role of CRISPR-Cas systems and anti-CRISPR proteins in bacterial antibiotic resistance
- PMID: 39149034
- PMCID: PMC11325803
- DOI: 10.1016/j.heliyon.2024.e34692
Role of CRISPR-Cas systems and anti-CRISPR proteins in bacterial antibiotic resistance
Abstract
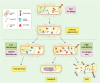
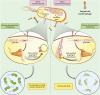
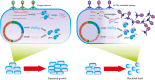
The emergence and development of antibiotic resistance in bacteria is a serious threat to global public health. Antibiotic resistance genes (ARGs) are often located on mobile genetic elements (MGEs). They can be transferred among bacteria by horizontal gene transfer (HGT), leading to the spread of drug-resistant strains and antibiotic treatment failure. CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated genes) is one of the many strategies bacteria have developed under long-term selection pressure to restrict the HGT. CRISPR-Cas systems exist in about half of bacterial genomes and play a significant role in limiting the spread of antibiotic resistance. On the other hand, bacteriophages and other MGEs encode a wide range of anti-CRISPR proteins (Acrs) to counteract the immunity of the CRISPR-Cas system. The Acrs could decrease the CRISPR-Cas system's activity against phages and facilitate the acquisition of ARGs and virulence traits for bacteria. This review aimed to assess the relationship between the CRISPR-Cas systems and Acrs with bacterial antibiotic resistance. We also highlighted the CRISPR technology and Acrs to control and prevent antibacterial resistance. The CRISPR-Cas system can target nucleic acid sequences with high accuracy and reliability; therefore, it has become a novel gene editing and gene therapy tool to prevent the spread of antibiotic resistance. CRISPR-based approaches may pave the way for developing smart antibiotics, which could eliminate multidrug-resistant (MDR) bacteria and distinguish between pathogenic and beneficial microorganisms. Additionally, the engineered anti-CRISPR gene-containing phages in combination with antibiotics could be used as a cutting-edge treatment approach to reduce antibiotic resistance.
Keywords: Anti-CRISPR proteins; Antibiotic resistance genes; CRISPR-Cas system; Horizontal gene transfer; Mobile genetic elements.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Soucy S.M., Huang J., Gogarten J.P. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 2015;16(8):472–482. - PubMed
-
- Frost L.S., Leplae R., Summers A.O., Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 2005 Sep;3(9):722–732. - PubMed
-
- Kaplan T. The role of horizontal gene transfer in antibiotic resistance. Eukaryon. 2014;10:80–81.
-
- Baltrus D.A. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 2013;28(8):489–495. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

