Targeted suppression of oral squamous cell carcinoma by pyrimidine-tethered quinoxaline derivatives
- PMID: 39149105
- PMCID: PMC11324040
- DOI: 10.1039/d4md00042k
Targeted suppression of oral squamous cell carcinoma by pyrimidine-tethered quinoxaline derivatives
Abstract
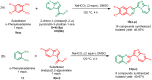
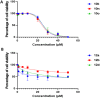
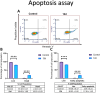
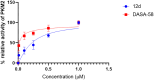
Oral cancer (OC) stands as a prominent cause of global mortality. Despite numerous efforts in recent decades, the efficacy of novel therapies to extend the lifespan of OC patients remains disappointingly low. Consequently, the demand for innovative therapeutic agents has become all the more pressing. In this context, we present our work on the design and synthesis of twenty-five novel quinoxaline-tethered imidazopyri(mi)dine derivatives. This was followed by comprehensive investigations into the impact of these molecules on the OC cell line. The in vitro cytotoxicity studies performed in CAL-27 and normal oral epithelial (NOE) cell lines revealed that some of the synthesized molecules like 12d have potent antiproliferative activity specifically towards OC cells with an IC50 of 0.79 μM and show negligible cytotoxicity over NOE cells. Further, 12d arrested cell growth in the S phase of the cell cycle and induced cell death by early apoptosis. The in silico studies validated that 12d binds to the activator binding site on pyruvate kinase M2 (PKM2) overexpressed in OC while the lactate dehydrogenase (LDH)-coupled enzyme assay established 12d as a potent PKM2 activator with an AC50 of 0.6 nM. Hence, this study provides fruitful evidence for the designed compounds as anticancer agents against OC.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors have no competing financial interests.
Figures









Similar articles
-
Novel imidazopyrimidines-based molecules induce tetramerization of tumor pyruvate kinase M2 and exhibit potent antiproliferative profile.Eur J Pharm Sci. 2022 Mar 1;170:106112. doi: 10.1016/j.ejps.2021.106112. Epub 2021 Dec 28. Eur J Pharm Sci. 2022. PMID: 34971746
-
Design, synthesis, and biological evaluation of 6-(imidazo[1,2-a] pyridin-6-yl) quinazolin-4(3H)-one derivatives as potent anticancer agents by dual targeting Aurora kinase and ROR1.Bioorg Chem. 2023 Jun;135:106484. doi: 10.1016/j.bioorg.2023.106484. Epub 2023 Mar 20. Bioorg Chem. 2023. PMID: 36963371
-
Novel halogenated arylvinyl-1,2,4 trioxanes as potent antiplasmodial as well as anticancer agents: Synthesis, bioevaluation, structure-activity relationship and in-silico studies.Eur J Med Chem. 2021 Nov 15;224:113685. doi: 10.1016/j.ejmech.2021.113685. Epub 2021 Jul 10. Eur J Med Chem. 2021. PMID: 34303874
-
Anti-tumor effects of novel alkannin derivatives with potent selectivity on comprehensive analysis.Phytomedicine. 2023 Aug;117:154912. doi: 10.1016/j.phymed.2023.154912. Epub 2023 Jun 2. Phytomedicine. 2023. PMID: 37295023
-
Development of novel tetrazole-based pyruvate kinase M2 inhibitors targeting U87MG glioblastoma cells.Bioorg Chem. 2025 Jan;154:108029. doi: 10.1016/j.bioorg.2024.108029. Epub 2024 Dec 5. Bioorg Chem. 2025. PMID: 39693922
References
-
- Oral Cancer, https://www.who.int/news-room/fact-sheets/detail/oral-health, (accessed on 12th May 2024)
-
- Mouth and oral cancer statistics, https://www.wcrf.org/cancer-trends/mouth-and-oral-cancer-statistics, (accessed on 12th May 2024)
LinkOut - more resources
Full Text Sources
Miscellaneous

