This is a preprint.
ISGylation of the SARS-CoV-2 N protein by HERC5 impedes N oligomerization and thereby viral RNA synthesis
- PMID: 39149229
- PMCID: PMC11326284
- DOI: 10.1101/2024.05.15.594393
ISGylation of the SARS-CoV-2 N protein by HERC5 impedes N oligomerization and thereby viral RNA synthesis
Update in
-
ISGylation of the SARS-CoV-2 N protein by HERC5 impedes N oligomerization and thereby viral RNA synthesis.J Virol. 2024 Sep 17;98(9):e0086924. doi: 10.1128/jvi.00869-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194248 Free PMC article.
Abstract
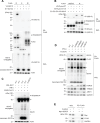
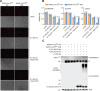
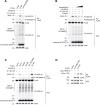
Interferon (IFN)-stimulated gene 15 (ISG15), a ubiquitin-like protein, is covalently conjugated to host (immune) proteins such as MDA5 and IRF3 in a process called ISGylation, thereby limiting the replication of Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, whether SARS-CoV-2 proteins can be directly targeted for ISGylation remains elusive. In this study, we identified the nucleocapsid (N) protein of SARS-CoV-2 as a major substrate of ISGylation catalyzed by the host E3 ligase HERC5; however, N ISGylation is readily removed through de-ISGylation by the papain-like protease (PLpro) activity of NSP3. Mass spectrometry analysis identified that the N protein undergoes ISGylation at four lysine residues (K266, K355, K387 and K388), and mutational analysis of these sites in the context of a SARS-CoV-2 replicon (N-4KR) abolished N ISGylation and alleviated ISGylation-mediated inhibition of viral RNA synthesis. Furthermore, our results indicated that HERC5 targets preferentially phosphorylated N protein for ISGylation to regulate its oligomeric assembly. These findings reveal a novel mechanism by which the host ISGylation machinery directly targets SARS-CoV-2 proteins to restrict viral replication and illuminate how an intricate interplay of host (HERC5) and viral (PLpro) enzymes coordinates viral protein ISGylation and thereby regulates virus replication.
Keywords: HERC5; ISG15; ISGylation; N protein; SARS-CoV-2; papain-like protease.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
