This is a preprint.
dia-PASEF Proteomics of Tumor and Stroma LMD Enriched from Archived HNSCC Samples
- PMID: 39149249
- PMCID: PMC11326218
- DOI: 10.1101/2024.08.09.607341
dia-PASEF Proteomics of Tumor and Stroma LMD Enriched from Archived HNSCC Samples
Update in
-
dia-PASEF Proteomics of Tumor and Stroma LMD Enriched from Archived HNSCC Samples.ACS Omega. 2025 Mar 25;10(13):13296-13302. doi: 10.1021/acsomega.4c11051. eCollection 2025 Apr 8. ACS Omega. 2025. PMID: 40224439 Free PMC article.
Abstract
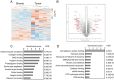
We employed laser microdissection to selectively harvest tumor cells and stroma from the microenvironment of formalin-fixed, paraffin-embedded head and neck squamous cell carcinoma (HNSCC) tissues. The captured HNSCC tissue fractions were analyzed by quantitative mass spectrometry-based proteomics using a data independent analysis approach. In paired samples, we achieved excellent proteome coverage having quantified 6,668 proteins with a median quantitative coefficient of variation under 10%. We observed significant differences in relevant functional pathways between the spatially resolved tumor and stroma regions. Our results identified extracellular matrix (ECM) as a major component enriched in the stroma, including many cancer associated fibroblast signature proteins in this compartment. We demonstrate the potential for comparative deep proteome analysis from very low starting input in a scalable format that is useful to decipher the alterations in tumor and the stromal microenvironment. Correlating such results with clinical features or disease progression will likely enable identification of novel targets for disease classification and interventions.
Conflict of interest statement
Conflict of interest AP, ALH, BVK, BD, TPC, RG declare that they have no known competing financial interests. DA, MW are employees of Bruker Scientific.
Figures


References
-
- Demichev V, Szyrwiel L, Yu F, Teo GC, Rosenberger G, Niewienda A, Ludwig D, Decker J, Kaspar-Schoenefeld S, Lilley KS, Mülleder M, Nesvizhskii AI, Ralser M. 2022. dia-PASEF data analysis using FragPipe and DIA-NN for deep proteomics of low sample amounts. Nat Commun 13:3944. doi: 10.1038/s41467-022-31492-0 - DOI - PMC - PubMed
-
- Huang C, Chen L, Savage SR, Eguez RV, Dou Y, Li Y, da Veiga Leprevost F, Jaehnig EJ, Lei JT, Wen B, Schnaubelt M, Krug K, Song X, Cieślik M, Chang H-Y, Wyczalkowski MA, Li K, Colaprico A, Li QK, Clark DJ, Hu Y, Cao L, Pan J, Wang Y, Cho K-C, Shi Z, Liao Y, Jiang W, Anurag M, Ji J, Yoo S, Zhou DC, Liang W-W, Wendl M, Vats P, Carr SA, Mani DR, Zhang Z, Qian J, Chen XS, Pico AR, Wang P, Chinnaiyan AM, Ketchum KA, Kinsinger CR, Robles AI, An E, Hiltke T, Mesri M, Thiagarajan M, Weaver AM, Sikora AG, Lubiński J, Wierzbicka M, Wiznerowicz M, Satpathy S, Gillette MA, Miles G, Ellis MJ, Omenn GS, Rodriguez H, Boja ES, Dhanasekaran SM, Ding L, Nesvizhskii AI, El-Naggar AK, Chan DW, Zhang H, Zhang B, Clinical Proteomic Tumor Analysis Consortium. 2021. Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer Cell 39:361–379.e16. doi: 10.1016/j.ccell.2020.12.007 - DOI - PMC - PubMed
-
- Hunt AL, Bateman NW, Barakat W, Makohon-Moore S, Hood BL, Conrads KA, Zhou M, Calvert V, Pierobon M, Loffredo J, Litzi TJ, Oliver J, Mitchell D, Gist G, Rojas C, Blanton B, Robinson EL, Odunsi K, Sood AK, Casablanca Y, Darcy KM, Shriver CD, Petricoin EF, Rao UNM, Maxwell GL, Conrads TP. 2021. Extensive three-dimensional intratumor proteomic heterogeneity revealed by multiregion sampling in high-grade serous ovarian tumor specimens. iScience 24:102757. doi: 10.1016/j.isci.2021.102757 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
