This is a preprint.
A Strain of Streptococcus mitis Inhibits Biofilm Formation of Caries Pathogens via Abundant Hydrogen Peroxide Production
- PMID: 39149263
- PMCID: PMC11326308
- DOI: 10.1101/2024.08.06.606862
A Strain of Streptococcus mitis Inhibits Biofilm Formation of Caries Pathogens via Abundant Hydrogen Peroxide Production
Update in
-
A strain of Streptococcus mitis inhibits biofilm formation of caries pathogens via abundant hydrogen peroxide production.Appl Environ Microbiol. 2025 Mar 19;91(3):e0219224. doi: 10.1128/aem.02192-24. Epub 2025 Feb 25. Appl Environ Microbiol. 2025. PMID: 39998256 Free PMC article.
Abstract
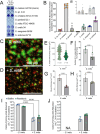
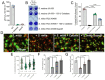
Commensal oral streptococci that colonize supragingival biofilms deploy mechanisms to combat competitors within their niche. Here, we determined that Streptococcus mitis more effectively inhibited biofilm formation of Streptococcus mutans within a seven species panel. This phenotype was common amongst all assayed isolates of S. mutans, but was specific to a single strain of S. mitis, ATCC 49456. The growth inhibitory factor was not effectively carried in spent supernatants of S. mitis. However, we documented ATCC 49456 to accumulate 4-5 times more hydrogen peroxide (H2O2) than other species tested, and 5-18 times more than other S. mitis strains assayed. The S. mutans biofilm formation inhibitory phenotype was reduced when grown in media containing catalase or with a S. mitis mutant of pyruvate oxidase (spxB; pox), confirming that SpxB-dependent H2O2 production was the main antagonistic factor. Addition of S. mitis within hours after S. mutans inoculation was effective at reducing biofilm biomass, but not for 24 h pre-formed biofilms. Transcriptome analysis revealed responses for both S. mitis and S. mutans, with several S. mutans differentially expressed genes following a gene expression pattern previously described, while others being unique to the interaction with S. mitis. Finally, we show that S. mitis also affected coculture biofilm formation of several other commensal streptococci. Our study shows that strains with abundant H2O2 production are effective at inhibiting initial growth of caries pathogens like S. mutans, but are less effective at disrupting pre-formed biofilms and have the potential to influence the stability of other oral commensal strains.
Conflict of interest statement
DECLARATION OF CONFLICTING INTERESTS The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures





References
-
- Peres MA, Macpherson LMD, Weyant RJ, Daly B, Venturelli R, Mathur MR, Listl S, Celeste RK, Guarnizo-Herreño CC, Kearns C, Benzian H, Allison P, Watt RG. 2019. Oral diseases: a global public health challenge. Lancet 394:249–260. - PubMed
-
- Ahmady K, Marsh PD, Newman HN, Bulman JS. 1993. Distribution of Streptococcus mutans and Streptococcus sobrinus at sub-sites in human approximal dental plaque. Caries Res 27:135–139. - PubMed
-
- Pitts NB, Zero DT, Marsh PD, Ekstrand K, Weintraub JA, Ramos-Gomez F, Tagami J, Twetman S, Tsakos G, Ismail A. 2017. Dental caries. Nat Rev Dis Prim 3:17030. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
