This is a preprint.
Identification of Meibomian gland stem cell populations and mechanisms of aging
- PMID: 39149265
- PMCID: PMC11326261
- DOI: 10.1101/2024.08.09.607015
Identification of Meibomian gland stem cell populations and mechanisms of aging
Update in
-
Identification of Meibomian gland stem cell populations and mechanisms of aging.Nat Commun. 2025 Feb 15;16(1):1663. doi: 10.1038/s41467-025-56907-6. Nat Commun. 2025. PMID: 39955307 Free PMC article.
Abstract
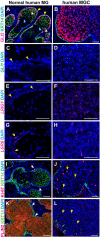
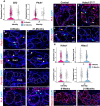
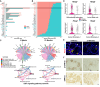
Meibomian glands secrete lipid-rich meibum, which prevents tear evaporation. Aging-related Meibomian gland shrinkage may result in part from stem cell exhaustion and is associated with evaporative dry eye disease, a common condition lacking effective treatment. The identities and niche of Meibomian gland stem cells and the signals controlling their activity are poorly defined. Using snRNA-seq, in vivo lineage tracing, ex vivo live imaging, and genetic studies in mice, we identified markers for stem cell populations that maintain distinct regions of the gland and uncovered Hh signaling as a key regulator of stem cell proliferation. Consistent with this, human Meibomian gland carcinoma exhibited increased Hh signaling. Aged glands displayed decreased Hh and EGF signaling, deficient innervation, and loss of collagen I in niche fibroblasts, indicating that alterations in both glandular epithelial cells and their surrounding microenvironment contribute to age-related degeneration. These findings suggest new approaches to treat aging-associated Meibomian gland loss.
Keywords: EGF; GLI2; Hedgehog signaling; Meibomian gland; Smoothened; lineage tracing; live imaging; snRNA-seq; stem cells.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Knop E., Knop N., Millar T., Obata H., and Sullivan D.A. (2011). The international workshop on meibomian gland dysfunction: report of the subcommittee on anatomy, physiology, and pathophysiology of the meibomian gland. Invest Ophthalmol Vis Sci 52, 1938–1978. 10.1167/iovs.10-6997c. - DOI - PMC - PubMed
-
- Wolff E., Bron A.J., Tripathi R.C., and Tripathi B.J. (1997). Anatomy of the eye and orbit, 8th Edition (Chapman & Hall Medical; ).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
