This is a preprint.
Improved vectors for retron-mediated CRISPR-Cas9 genome editing in Saccharomyces cerevisiae
- PMID: 39149293
- PMCID: PMC11326209
- DOI: 10.1101/2024.08.06.606807
Improved vectors for retron-mediated CRISPR-Cas9 genome editing in Saccharomyces cerevisiae
Update in
-
Improved vectors for retron-mediated CRISPR-Cas9 genome editing in Saccharomyces cerevisiae.G3 (Bethesda). 2025 Oct 8;15(10):jkaf175. doi: 10.1093/g3journal/jkaf175. G3 (Bethesda). 2025. PMID: 40758833 Free PMC article.
Abstract
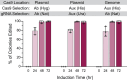
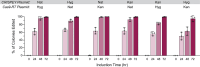
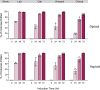
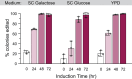
In vivo site-directed mutagenesis is a powerful genetic tool for testing the effects of specific alleles in their normal genomic context. While the budding yeast Saccharomyces cerevisiae possesses classical tools for site-directed mutagenesis, more efficient recent CRISPR-based approaches use Cas 'cutting' combined with homologous recombination of a 'repair' template that introduces the desired edit. However, current approaches are limited for fully prototrophic yeast strains, and rely on relatively low efficiency cloning of short gRNAs. We were thus motivated to simplify the process by combining the gRNA and its cognate repair template in cis on a single oligonucleotide. Moreover, we wished to take advantage of a new approach that uses an E. coli retron (EcRT) to amplify repair templates as multi-copy single-stranded (ms)DNA in vivo, which are more efficient templates for homologous recombination. To this end, we have created a set of plasmids that express Cas9-EcRT, allowing for co-transformation with the gRNA-repair template plasmid in a single step. Our suite of plasmids contains different antibiotic (Nat, Hyg, Kan) or auxotrophic (HIS3, URA3) selectable markers, allowing for editing of fully prototrophic wild yeast strains. In addition to classic galactose induction, we generated a β-estradiol-inducible version of each plasmid to facilitate editing in yeast strains that grow poorly on galactose. The plasmid-based system results in >95% editing efficiencies for point mutations and >50% efficiencies for markerless deletions, in a minimum number of steps and time. We provide a detailed step-by-step guide for how to use this system.
Figures

References
-
- Arita Y., Kim G., Li Z., Friesen H., Turco G., Wang R. Y., Climie D., Usaj M., Hotz M., Stoops E. H., Baryshnikova A., Boone C., Botstein D., Andrews B. J., & McIsaac R. S. (2021). A genome-scale yeast library with inducible expression of individual genes. Molecular Systems Biology, 17(6). - PMC - PubMed
-
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., & Boeke J. D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14(2), 115–132. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
