This is a preprint.
TFE3 fusions direct an oncogenic transcriptional program that drives OXPHOS and unveils vulnerabilities in translocation renal cell carcinoma
- PMID: 39149323
- PMCID: PMC11326252
- DOI: 10.1101/2024.08.09.607311
TFE3 fusions direct an oncogenic transcriptional program that drives OXPHOS and unveils vulnerabilities in translocation renal cell carcinoma
Update in
-
Oncogenic TFE3 fusions drive OXPHOS and confer metabolic vulnerabilities in translocation renal cell carcinoma.Nat Metab. 2025 Mar;7(3):478-492. doi: 10.1038/s42255-025-01218-9. Epub 2025 Feb 6. Nat Metab. 2025. PMID: 39915638
Abstract
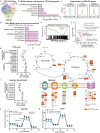
Translocation renal cell carcinoma (tRCC) is an aggressive subtype of kidney cancer driven by TFE3 gene fusions, which act via poorly characterized downstream mechanisms. Here we report that TFE3 fusions transcriptionally rewire tRCCs toward oxidative phosphorylation (OXPHOS), contrasting with the highly glycolytic metabolism of most other renal cancers. This TFE3 fusion-driven OXPHOS program, together with heightened glutathione levels found in renal cancers, renders tRCCs sensitive to reductive stress - a metabolic stress state induced by an imbalance of reducing equivalents. Genome-scale CRISPR screening identifies tRCC-selective vulnerabilities linked to this metabolic state, including EGLN1, which hydroxylates HIF-1α and targets it for proteolysis. Inhibition of EGLN1 compromises tRCC cell growth by stabilizing HIF-1a and promoting metabolic reprogramming away from OXPHOS, thus representing a vulnerability to OXPHOS-dependent tRCC cells. Our study defines a distinctive tRCC-essential metabolic program driven by TFE3 fusions and nominates EGLN1 inhibition as a therapeutic strategy to counteract fusion-induced metabolic rewiring.
Conflict of interest statement
Declaration of Interests S.R.V. has consulted for Jnana Therapeutics within the past 3 years and receives research support from Bayer. L.B-P. is a co-founder, holds equity in and is a consultant for Scorpion Therapeutics.
Figures


References
-
- Moch H., Cubilla A. L., Humphrey P. A., Reuter V. E. & Ulbright T. M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs—Part A: Renal, Penile, and Testicular Tumours. European Urology 70, 93–105 (2016). - PubMed
-
- Argani P. et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol 31, 1149–1160 (2007). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
