This is a preprint.
Sleep and diurnal alternative polyadenylation sites associated with human APA-linked brain disorders
- PMID: 39149473
- PMCID: PMC11326403
- DOI: 10.21203/rs.3.rs-4707772/v1
Sleep and diurnal alternative polyadenylation sites associated with human APA-linked brain disorders
Update in
-
Sleep and diurnal alternative polyadenylation sites associated with human APA-linked brain disorders.NPJ Biol Timing Sleep. 2024;1(1):11. doi: 10.1038/s44323-024-00012-2. Epub 2024 Nov 1. NPJ Biol Timing Sleep. 2024. PMID: 39493890 Free PMC article.
Abstract
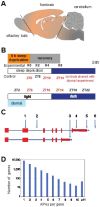
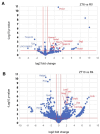
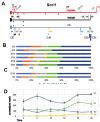
Disruption of sleep and circadian rhythms are a comorbid feature of many pathologies, and can negatively influence many health conditions, including neurodegenerative disease, metabolic illness, cancer, and various neurological disorders. Genetic association studies linking sleep and circadian disturbances with disease susceptibility have mainly focused on changes in gene expression due to mutations, such as single-nucleotide polymorphisms. The interaction between sleep and/or circadian rhythms with the use of Alternative Polyadenylation (APA) has been largely undescribed, particularly in the context of other disorders. APA is a process that generates various transcript isoforms of the same gene affecting its mRNA translation, stability, localization, and subsequent function. Here we identified unique APAs expressed in rat brain over time-of-day, immediately following sleep deprivation, and the subsequent recovery period. From these data, we performed a secondary analysis of these sleep- or time-of-day associated PASs with recently described APA-linked human brain disorder susceptibility genes.
Conflict of interest statement
Competing Interests J.G. is the founder of Blood Brain Biotechnology, LLC and a member of its scientific advisory board. Jon Davis is a principal scientist at Integrated Physiology Research, Novo Nordisk. All other authors declare no competing interests.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

