This is a preprint.
Structure-Informed Design of an Ultra Bright RNA-activated Fluorophore
- PMID: 39149476
- PMCID: PMC11326382
- DOI: 10.21203/rs.3.rs-4750449/v1
Structure-Informed Design of an Ultra Bright RNA-activated Fluorophore
Update in
-
Structure-informed design of an ultrabright RNA-activated fluorophore.Nat Chem. 2025 Aug;17(8):1188-1195. doi: 10.1038/s41557-025-01832-w. Epub 2025 May 28. Nat Chem. 2025. PMID: 40437193 Free PMC article.
Abstract
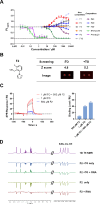
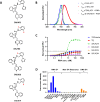
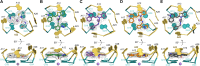
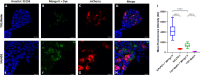
Fluorogenic RNAs such as the Mango aptamers are uniquely powerful tools for imaging RNA. A central challenge has been to develop brighter, more specific, and higher affinity aptamer-ligand systems for cellular imaging. Here, we report an ultra-bright fluorophore for the Mango II system discovered using a structure-informed, fragment-based small molecule microarray approach. The new dye, Structure informed, Array-enabled LigAnD 1 (SALAD1) exhibits 3.5-fold brighter fluorescence than TO1-Biotin and subnanomolar aptamer affinity. Improved performance comes solely from alteration of dye-RNA interactions, without alteration of the chromophore itself. Multiple high-resolution structures reveal a unique and specific binding mode for the new dye resulting from improved pocket occupancy, a more defined binding pose, and a novel bonding interaction with potassium. The dye notably improves in-cell confocal RNA imaging. This work provides both introduces a new RNA-activated fluorophore and also a powerful demonstration of how to leverage fragment-based ligand discovery against RNA targets.
Figures

References
-
- Yin P, Kuang S, Nie Z. Fluorescent RNA Tags for In Situ RNA Imaging in Living Cells. Analysis & Sensing 2023, 3(5): e202200090.
-
- Armitage BA. Imaging of RNA in live cells. Curr Opin Chem Biol 2011, 15(6): 806–812. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

