KREH2 helicase represses ND7 mRNA editing in procyclic-stage Trypanosoma brucei by opposite modulation of canonical and 'moonlighting' gRNA utilization creating a proposed mRNA structure
- PMID: 39149912
- PMCID: PMC11514453
- DOI: 10.1093/nar/gkae699
KREH2 helicase represses ND7 mRNA editing in procyclic-stage Trypanosoma brucei by opposite modulation of canonical and 'moonlighting' gRNA utilization creating a proposed mRNA structure
Abstract
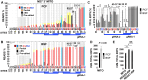
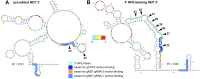
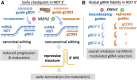
Unknown factors regulate mitochondrial U-insertion/deletion (U-indel) RNA editing in procyclic-form (PCF) and bloodstream-form (BSF) T. brucei. This editing, directed by anti-sense gRNAs, creates canonical protein-encoding mRNAs and may developmentally control respiration. Canonical editing by gRNAs that specify protein-encoding mRNA sequences occurs amid massive non-canonical editing of unclear sources and biological significance. We found PCF-specific repression at a major early checkpoint in mRNA ND7, involving helicase KREH2-dependent opposite modulation of canonical and non-canonical 'terminator' gRNA utilization. Terminator-programmed editing derails canonical editing and installs proposed repressive structure in 30% of the ND7 transcriptome. BSF-to-PCF differentiation in vitro recreated this negative control. Remarkably, KREH2-RNAi knockdown relieved repression and increased editing progression by reverting canonical/terminator gRNA utilization. ND7 transcripts lacking early terminator-directed editing in PCF exhibited similar negative editing control along the mRNA sequence, suggesting global modulation of gRNA utilization fidelity. The terminator is a 'moonlighting' gRNA also associated with mRNA COX3 canonical editing, so the gRNA transcriptome seems multifunctional. Thus, KREH2 is the first identified repressor in developmental editing control. This and our prior work support a model whereby KREH2 activates or represses editing in a stage and substrate-specific manner. KREH2's novel dual role tunes mitochondrial gene expression in either direction during development.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures








References
-
- Zimmer S.L. Revisiting trypanosome mitochondrial genome mysteries: broader and deeper. Trends Parasitol. 2019; 35:102–104. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

