Destabilization of the D2 domain of Thermotoga maritima arginine binding protein induced by guanidinium thiocyanate and its counteraction by stabilizing agents
- PMID: 39150147
- PMCID: PMC11328109
- DOI: 10.1002/pro.5146
Destabilization of the D2 domain of Thermotoga maritima arginine binding protein induced by guanidinium thiocyanate and its counteraction by stabilizing agents
Abstract
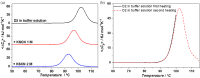
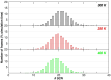
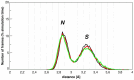
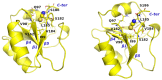
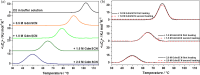
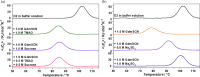
D2 is a structural and cooperative domain of Thermotoga maritima Arginine Binding Protein, that possesses a remarkable conformational stability, with a denaturation temperature of 102.6°C, at pH 7.4. The addition of potassium thiocyanate causes a significant decrease in the D2 denaturation temperature. The interactions of thiocyanate ions with D2 have been studied by means of isothermal titration calorimetry measurements and molecular dynamics simulations. It emerged that: (a) 20-30 thiocyanate ions interact with the D2 surface and are present in its first solvation shell; (b) each of them makes several contacts with protein groups, both polar and nonpolar ones. The addition of guanidinium thiocyanate causes a marked destabilization of the D2 native state, because both the ions are denaturing agents. However, on adding to the solution containing D2 and guanidinium thiocyanate a stabilizing agent, such as TMAO, sucrose or sodium sulfate, a significant increase in denaturation temperature occurs. The present results confirm that counteraction is a general phenomenon for globular proteins.
Keywords: Guanidinium; MD simulations; TmArgBP; calorimetry; thermodynamics; thiocyanate.
© 2024 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures


References
-
- Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J Phys Chem. 1987;91:6269–6271. 10.1021/j100308a038 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

