Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis
- PMID: 39150221
- PMCID: PMC11328120
- DOI: 10.1002/pro.5144
Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis
Abstract
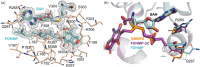
MpaG' is an S-adenosyl-L-methionine (SAM)-dependent methyltransferase involved in the compartmentalized biosynthesis of mycophenolic acid (MPA), a first-line immunosuppressive drug for organ transplantations and autoimmune diseases. MpaG' catalyzes the 5-O-methylation of three precursors in MPA biosynthesis including demethylmycophenolic acid (DMMPA), 4-farnesyl-3,5-dihydroxy-6-methylphthalide (FDHMP), and an intermediate containing three fewer carbon atoms compared to FDHMP (FDHMP-3C) with different catalytic efficiencies. Here, we report the crystal structures of S-adenosyl-L-homocysteine (SAH)/DMMPA-bound MpaG', SAH/FDHMP-3C-bound MpaG', and SAH/FDHMP-bound MpaG' to understand the catalytic mechanism of MpaG' and structural basis for its substrate flexibility. Structural and biochemical analyses reveal that MpaG' utilizes the catalytic dyad H306-E362 to deprotonate the C5 hydroxyl group of the substrates for the following methylation. The three substrates with differently modified farnesyl moieties are well accommodated in a large semi-open substrate binding pocket with the orientation of their phthalide moiety almost identical. Based on the structure-directed mutagenesis, a single mutant MpaG'Q267A is engineered with significantly improved catalytic efficiency for all three substrates. This study expands the mechanistic understanding and the pocket engineering strategy for O-methyltransferases involved in fungal natural product biosynthesis. Our research also highlights the potential of O-methyltransferases to modify diverse substrates by protein design and engineering.
Keywords: O‐methyltransferase; crystal structure; mycophenolic acid; protein engineering; substrate flexibility.
© 2024 The Protein Society.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
MeSH terms
Substances
Grants and funding
- 2023YFC3402300/National Key Research and Development Program of China
- 22237004/National Natural Science Foundation of China
- 32200030/National Natural Science Foundation of China
- 32070125/National Natural Science Foundation of China
- M2022-01/State Key Laboratory of Microbial Technology Open Projects Fund
LinkOut - more resources
Full Text Sources
Miscellaneous

