From raw milk cheese to the gut: investigating the colonization strategies of Bifidobacterium mongoliense
- PMID: 39150265
- PMCID: PMC11409640
- DOI: 10.1128/aem.01244-24
From raw milk cheese to the gut: investigating the colonization strategies of Bifidobacterium mongoliense
Abstract
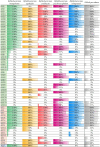
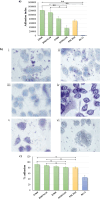
The microbial ecology of raw milk cheeses is determined by bacteria originating from milk and milk-producing animals. Recently, it has been shown that members of the Bifidobacterium mongoliense species may become transmitted along the Parmigiano Reggiano cheese production chain and ultimately may colonize the consumer intestine. However, there is a lack of knowledge regarding the molecular mechanisms that mediate the interaction between B. mongoliense and the human gut. Based on 128 raw milk cheeses collected from different Italian regions, we isolated and characterized 10 B. mongoliense strains. Comparative genomics allowed us to unveil the presence of enzymes required for the degradation of sialylated host-glycans in B. mongoliense, corroborating the appreciable growth on de Man-Rogosa-Sharpe (MRS) medium supplemented with 3'-sialyllactose (3'-SL) or 6'-sialyllactose (6'-SL). The B. mongoliense BMONG18 was chosen, due to its superior ability to utilize 3'-SL and mucin as representative strain, to investigate its behavior when co-inoculated with other bifidobacterial species. Conversely, members of other bifidobacterial species did not appear to benefit from the presence of BMONG18, highlighting a competitive scenario for nutrient acquisition. Transcriptomic data of BMONG18 reveal no significant differences in gene expression when cultivated in a gut simulating medium (GSM), regardless of whether cheese was included or not. Furthermore, BMONG18 was shown to exhibit high adhesion capabilities to HT29-MTX human cells, in line with its colonization ability of a human host.IMPORTANCEFermented foods are nourishments produced through controlled microbial growth that play an essential role in worldwide human nutrition. Research interest in fermented foods has increased since the 80s, driven by growing awareness of their potential health benefits beyond mere nutritional content. Bifidobacterium mongoliense, previously identified throughout the production process of Parmigiano Reggiano cheese, was found to be capable of establishing itself in the intestines of its consumers. Our study underscores molecular mechanisms through which this bifidobacterial species, derived from food, interacts with the host and other gut microbiota members.
Keywords: bifidobacteria; dairy food microorganisms; microbe–host interactions; microbial genomics; microbiome; microbiota.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Detection and characterization of Bifidobacterium crudilactis and B. mongoliense able to grow during the manufacturing process of French raw milk cheeses.BMC Microbiol. 2013 Oct 29;13:239. doi: 10.1186/1471-2180-13-239. BMC Microbiol. 2013. PMID: 24164698 Free PMC article.
-
Bifidobacterium mongoliense genome seems particularly adapted to milk oligosaccharide digestion leading to production of antivirulent metabolites.BMC Microbiol. 2020 May 7;20(1):111. doi: 10.1186/s12866-020-01804-9. BMC Microbiol. 2020. PMID: 32380943 Free PMC article.
-
Colonization of the human gut by bovine bacteria present in Parmesan cheese.Nat Commun. 2019 Mar 20;10(1):1286. doi: 10.1038/s41467-019-09303-w. Nat Commun. 2019. PMID: 30894548 Free PMC article.
-
Invited review: Microbial evolution in raw-milk, long-ripened cheeses produced using undefined natural whey starters.J Dairy Sci. 2014 Feb;97(2):573-91. doi: 10.3168/jds.2013-7187. Epub 2013 Dec 2. J Dairy Sci. 2014. PMID: 24290824 Review.
-
The role of mucin and oligosaccharides via cross-feeding activities by Bifidobacterium: A review.Int J Biol Macromol. 2021 Jan 15;167:1329-1337. doi: 10.1016/j.ijbiomac.2020.11.087. Epub 2020 Nov 14. Int J Biol Macromol. 2021. PMID: 33202267 Review.
Cited by
-
Vitamin biosynthesis in the gut: interplay between mammalian host and its resident microbiota.Microbiol Mol Biol Rev. 2025 Jun 25;89(2):e0018423. doi: 10.1128/mmbr.00184-23. Epub 2025 Apr 2. Microbiol Mol Biol Rev. 2025. PMID: 40172109 Review.
-
Functional modulation of the human gut microbiome by bacteria vehicled by cheese.Appl Environ Microbiol. 2025 Mar 19;91(3):e0018025. doi: 10.1128/aem.00180-25. Epub 2025 Feb 28. Appl Environ Microbiol. 2025. PMID: 40019271 Free PMC article.
References
-
- Marco ML, Sanders ME, Gänzle M, Arrieta MC, Cotter PD, De Vuyst L, Hill C, Holzapfel W, Lebeer S, Merenstein D, Reid G, Wolfe BE, Hutkins R. 2021. The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on fermented foods. Nat Rev Gastroenterol Hepatol 18:196–208. doi:10.1038/s41575-020-00390-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

