Adaptive evolution of carbapenem-resistant hypervirulent Klebsiella pneumoniae in the urinary tract of a single patient
- PMID: 39150777
- PMCID: PMC11363291
- DOI: 10.1073/pnas.2400446121
Adaptive evolution of carbapenem-resistant hypervirulent Klebsiella pneumoniae in the urinary tract of a single patient
Abstract
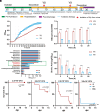
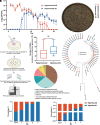
The emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKp) is a growing concern due to its high mortality and limited treatment options. Although hypermucoviscosity is crucial for CR-hvKp infection, the role of changes in bacterial mucoviscosity in the host colonization and persistence of CR-hvKp is not clearly defined. Herein, we observed a phenotypic switch of CR-hvKp from a hypermucoviscous to a hypomucoviscous state in a patient with scrotal abscess and urinary tract infection (UTI). This switch was attributed to decreased expression of rmpADC, the regulator of mucoid phenotype, caused by deletion of the upstream insertion sequence ISKpn26. Postswitching, the hypomucoid variant showed a 9.0-fold decrease in mice sepsis mortality, a >170.0-fold reduction in the ability to evade macrophage phagocytosis in vitro, and an 11.2- to 40.9-fold drop in growth rate in normal mouse serum. Conversely, it exhibited an increased residence time in the mouse urinary tract (21 vs. 6 d), as well as a 216.4-fold boost in adhesion to bladder epithelial cells and a 48.7% enhancement in biofilm production. Notably, the CR-hvKp mucoid switch was reproduced in an antibiotic-free mouse UTI model. The in vivo generation of hypomucoid variants was primarily associated with defective or low expression of rmpADC or capsule synthesis gene wcaJ, mediated by ISKpn26 insertion/deletion or base-pair insertion. The spontaneous hypomucoid variants also outcompeted hypermucoid bacteria in the mouse urinary tract. Collectively, the ISKpn26-associated mucoid switch in CR-hvKp signifies the antibiotic-independent host adaptive evolution, providing insights into the role of mucoid switch in the persistence of CR-hvKp.
Keywords: CR-hvKp; antibiotic-independent evolution; mucoid switch; persistence; urinary tract.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures





References
-
- Yang W., et al. , Current status and trends of antimicrobial resistance among clinical isolates in China: A retrospective study of CHINET from 2018 to 2022. One Health Adv. 1, 8 (2023).
-
- Gu D., et al. , A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: A molecular epidemiological study. Lancet Infect. Dis. 18, 37–46 (2018). - PubMed
-
- Wu Y., et al. , Global evolution and geographic diversity of hypervirulent carbapenem-resistant Klebsiella pneumoniae. Lancet Infect. Dis. 22, 761–762 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

