Light regulates widespread plant alternative polyadenylation through the chloroplast
- PMID: 39150783
- PMCID: PMC11348263
- DOI: 10.1073/pnas.2405632121
Light regulates widespread plant alternative polyadenylation through the chloroplast
Abstract
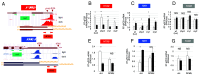
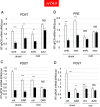
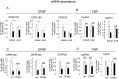
Transcription of eukaryotic protein-coding genes generates immature mRNAs that are subjected to a series of processing events, including capping, splicing, cleavage, and polyadenylation (CPA), and chemical modifications of bases. Alternative polyadenylation (APA) greatly contributes to mRNA diversity in the cell. By determining the length of the 3' untranslated region, APA generates transcripts with different regulatory elements, such as miRNA and RBP binding sites, which can influence mRNA stability, turnover, and translation. In the model plant Arabidopsis thaliana, APA is involved in the control of seed dormancy and flowering. In view of the physiological importance of APA in plants, we decided to investigate the effects of light/dark conditions and compare the underlying mechanisms to those elucidated for alternative splicing (AS). We found that light controls APA in approximately 30% of Arabidopsis genes. Similar to AS, the effect of light on APA requires functional chloroplasts, is not affected in mutants of the phytochrome and cryptochrome photoreceptor pathways, and is observed in roots only when the communication with the photosynthetic tissues is not interrupted. Furthermore, mitochondrial and TOR kinase activities are necessary for the effect of light. However, unlike AS, coupling with transcriptional elongation does not seem to be involved since light-dependent APA regulation is neither abolished in mutants of the TFIIS transcript elongation factor nor universally affected by chromatin relaxation caused by histone deacetylase inhibition. Instead, regulation seems to correlate with changes in the abundance of constitutive CPA factors, also mediated by the chloroplast.
Keywords: Arabidopsis thaliana; alternative polyadenylation; light control in plants.
Conflict of interest statement
Competing interests statement:The corresponding author of this report and one of the reviewers were co-authors of a 18-author review published online in 2022 (https://academic.oup.com/plcell/article/35/6/1626/6883923?login=false) in which none of them were corresponding authors.
Figures



References
-
- Liu F., Marquardt S., Lister C., Swiezewski S., Dean C., Targeted 3’ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97 (2010). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

