Functional overlap of inborn errors of immunity and metabolism genes defines T cell metabolic vulnerabilities
- PMID: 39151020
- PMCID: PMC11590014
- DOI: 10.1126/sciimmunol.adh0368
Functional overlap of inborn errors of immunity and metabolism genes defines T cell metabolic vulnerabilities
Abstract
Inborn errors of metabolism (IEMs) and immunity (IEIs) are Mendelian diseases in which complex phenotypes and patient rarity have limited clinical understanding. Whereas few genes have been annotated as contributing to both IEMs and IEIs, immunometabolic demands suggested greater functional overlap. Here, CRISPR screens tested IEM genes for immunologic roles and IEI genes for metabolic effects and found considerable previously unappreciated crossover. Analysis of IEMs showed that N-linked glycosylation and the hexosamine pathway enzyme Gfpt1 are critical for T cell expansion and function. Further, T helper (TH1) cells synthesized uridine diphosphate N-acetylglucosamine more rapidly and were more impaired by Gfpt1 deficiency than TH17 cells. Screening IEI genes found that Bcl11b promotes the CD4 T cell mitochondrial activity and Mcl1 expression necessary to prevent metabolic stress. Thus, a high degree of functional overlap exists between IEM and IEI genes, and immunometabolic mechanisms may underlie a previously underappreciated intersection of these disorders.
Conflict of interest statement
Figures


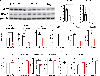


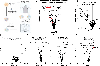
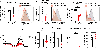

Update of
-
Functional Overlap of Inborn Errors of Immunity and Metabolism Genes Define T Cell Immunometabolic Vulnerabilities.bioRxiv [Preprint]. 2023 Jan 24:2023.01.24.525419. doi: 10.1101/2023.01.24.525419. bioRxiv. 2023. Update in: Sci Immunol. 2024 Aug 16;9(98):eadh0368. doi: 10.1126/sciimmunol.adh0368. PMID: 36747715 Free PMC article. Updated. Preprint.
References
-
- Sugiura A, Andrejeva G, Voss K, Heintzman DR, Xu X, Madden MZ, Ye X, Beier KL, Chowdhury NU, Wolf MM, Young AC, Greenwood DL, Sewell AE, Shahi SK, Freedman SN, Cameron AM, Foerch P, Bourne T, Garcia-Canaveras JC, Karijolich J, Newcomb DC, Mangalam AK, Rabinowitz JD, Rathmell JC, MTHFD2 is a metabolic checkpoint controlling effector and regulatory T cell fate and function. Immunity 55, 65–81 e69 (2022). - PMC - PubMed
-
- Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, Maseda D, Liberti MV, Paz K, Kishton RJ, Johnson ME, de Cubas AA, Wu P, Li G, Zhang Y, Newcomb DC, Wells AD, Restifo NP, Rathmell WK, Locasale JW, Davila ML, Blazar BR, Rathmell JC, Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175, 1780–1795 e19 (2018). - PMC - PubMed
-
- Gerriets VA, Kishton RJ, Nichols AG, Macintyre AN, Inoue M, Ilkayeva O, Winter PS, Liu X, Priyadharshini B, Slawinska ME, Haeberli L, Huck C, Turka LA, Wood KC, Hale LP, Smith PA, Schneider MA, MacIver NJ, Locasale JW, Newgard CB, Shinohara ML, Rathmell JC, Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125, 194–207 (2015). - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- T32 CA009582/CA/NCI NIH HHS/United States
- RF1 MH123971/MH/NIMH NIH HHS/United States
- R01 AI165722/AI/NIAID NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01 AI153167/AI/NIAID NIH HHS/United States
- S10 OD021630/OD/NIH HHS/United States
- R35 GM128915/GM/NIGMS NIH HHS/United States
- F99 NS125829/NS/NINDS NIH HHS/United States
- T32 AR059039/AR/NIAMS NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R35 GM155339/GM/NIGMS NIH HHS/United States
- R01 HL136664/HL/NHLBI NIH HHS/United States
- U01 AI176244/AI/NIAID NIH HHS/United States
- R01 DK105550/DK/NIDDK NIH HHS/United States
- R01 CA217987/CA/NCI NIH HHS/United States
- S10 OD012324/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

