The lipidomics reporting checklist a framework for transparency of lipidomic experiments and repurposing resource data
- PMID: 39151590
- PMCID: PMC11417233
- DOI: 10.1016/j.jlr.2024.100621
The lipidomics reporting checklist a framework for transparency of lipidomic experiments and repurposing resource data
Abstract
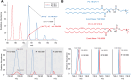
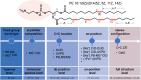
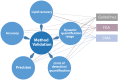
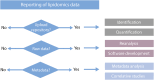
The rapid increase in lipidomic studies has led to a collaborative effort within the community to establish standards and criteria for producing, documenting, and disseminating data. Creating a dynamic easy-to-use checklist that condenses key information about lipidomic experiments into common terminology will enhance the field's consistency, comparability, and repeatability. Here, we describe the structure and rationale of the established Lipidomics Minimal Reporting Checklist to increase transparency in lipidomics research.
Keywords: FAIR; checklist; lipid metabolism; lipidomics; mass spectrometry; metabolomics; quality control; reference standards.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interests Kaddurah-Daouk is an inventor on key patents in the field of metabolomics and hold equity in Metabolon, a biotech company in North Carolina. In addition, she holds patents licensed to Chymia LLC and PsyProtix with royalties and ownership. Kim Ekroos is the owner of Lipidomics Consulting Ltd.
Figures





References
-
- Liebisch G., Ahrends R., Arita M., Arita M., Bowden J.A., Ejsing C.S., et al. Lipidomics needs more standardization. Nat. Metab. 2019;1:745–747. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

