Allele-specific transcription factor binding across human brain regions offers mechanistic insight into eQTLs
- PMID: 39152038
- PMCID: PMC11444172
- DOI: 10.1101/gr.278601.123
Allele-specific transcription factor binding across human brain regions offers mechanistic insight into eQTLs
Abstract
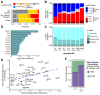
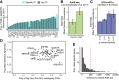
Transcription factors (TFs) regulate gene expression by facilitating or disrupting the formation of transcription initiation machinery at particular genomic loci. Because TF occupancy is driven in part by recognition of DNA sequence, genetic variation can influence TF-DNA associations and gene regulation. To identify variants that impact TF binding in human brain tissues, we assessed allele-specific binding (ASB) at heterozygous variants for 94 TFs in nine brain regions from two donors. Leveraging graph genomes constructed from phased genomic sequence data, we compared ChIP-seq signals between alleles at heterozygous variants within each brain region and identified thousands of variants exhibiting ASB for at least one TF. ASB reproducibility was measured by comparisons between independent experiments both within and between donors. We found that rare alleles in the general population more frequently led to reduced TF binding, whereas common alleles had an equal likelihood of increasing or decreasing binding. Further, for ASB variants in predicted binding motifs, the favored allele tended to be the one with the stronger expected motif match, but this concordance was not observed within highly occupied sites. We also found that neuron-specific cis-regulatory elements (cCREs), in contrast with oligodendrocyte-specific cCREs, showed depletion of ASB variants. We identified 2670 ASB variants associated with evidence for allele-specific gene expression in the brain from GTEx data and observed increasing eQTL effect direction concordance as ASB significance increases. These results provide a valuable and unique resource for mechanistic analysis of cis-regulatory variation in human brain tissue.
© 2024 Anderson et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Update of
-
Allele biased transcription factor binding across human brain regions gives mechanistic insight into eQTLs.bioRxiv [Preprint]. 2023 Oct 9:2023.10.06.561245. doi: 10.1101/2023.10.06.561245. bioRxiv. 2023. Update in: Genome Res. 2024 Sep 20;34(8):1224-1234. doi: 10.1101/gr.278601.123. PMID: 37873117 Free PMC article. Updated. Preprint.
References
-
- Anderson AG, Rogers BB, Loupe JM, Rodriguez-Nunez I, Roberts SC, White LM, Brazell JN, Bunney WE, Bunney BG, Watson SJ, et al. 2023. Single nucleus multiomics identifies ZEB1 and MAFB as candidate regulators of Alzheimer's disease-specific cis-regulatory elements. Cell Genomics 3: 100263. 10.1016/j.xgen.2023.100263 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
