Optimizing dsRNA engineering strategies and production in E. coli HT115 (DE3)
- PMID: 39152090
- PMCID: PMC11375590
- DOI: 10.1093/jimb/kuae028
Optimizing dsRNA engineering strategies and production in E. coli HT115 (DE3)
Abstract
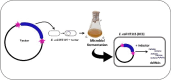
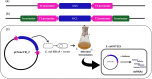
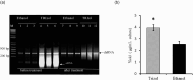
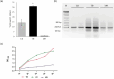
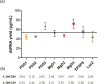
Producing double-stranded RNA (dsRNA) represents a bottleneck for the adoption of RNA interference technology in agriculture, and the main hurdles are related to increases in dsRNA yield, production efficiency, and purity. Therefore, this study aimed to optimize dsRNA production in E. coli HT115 (DE3) using an in vivo system. To this end, we designed a new vector, pCloneVR_2, which resulted in the efficient production of dsRNA in E. coli HT115 (DE3). We performed optimizations in the culture medium and expression inducer in the fermentation of E. coli HT115 (DE3) for the production of dsRNA. Notably, the variable that had the greatest effect on dsRNA yield was cultivation in TB medium, which resulted in a 118% increase in yield. Furthermore, lactose induction (6 g/L) yielded 10 times more than IPTG. Additionally, our optimized up-scaled protocol of the TRIzol™ extraction method was efficient for obtaining high-quality and pure dsRNA. Finally, our optimized protocol achieved an average yield of 53.3 µg/mL after the production and purification of different dsRNAs, reducing production costs by 72%.
Keywords: Bacterial dsRNA production; Double-stranded RNA; Lactose induction; RNAi.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society of Industrial Microbiology and Biotechnology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

