De novo design of miniprotein antagonists of cytokine storm inducers
- PMID: 39152100
- PMCID: PMC11329760
- DOI: 10.1038/s41467-024-50919-4
De novo design of miniprotein antagonists of cytokine storm inducers
Abstract
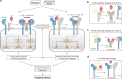
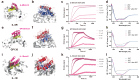
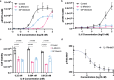
Cytokine release syndrome (CRS), commonly known as cytokine storm, is an acute systemic inflammatory response that is a significant global health threat. Interleukin-6 (IL-6) and interleukin-1 (IL-1) are key pro-inflammatory cytokines involved in CRS and are hence critical therapeutic targets. Current antagonists, such as tocilizumab and anakinra, target IL-6R/IL-1R but have limitations due to their long half-life and systemic anti-inflammatory effects, making them less suitable for acute or localized treatments. Here we present the de novo design of small protein antagonists that prevent IL-1 and IL-6 from interacting with their receptors to activate signaling. The designed proteins bind to the IL-6R, GP130 (an IL-6 co-receptor), and IL-1R1 receptor subunits with binding affinities in the picomolar to low-nanomolar range. X-ray crystallography studies reveal that the structures of these antagonists closely match their computational design models. In a human cardiac organoid disease model, the IL-1R antagonists demonstrated protective effects against inflammation and cardiac damage induced by IL-1β. These minibinders show promise for administration via subcutaneous injection or intranasal/inhaled routes to mitigate acute cytokine storm effects.
© 2024. The Author(s).
Conflict of interest statement
B.H., B.C., M. Abedi., L.C., L.S., and D.B. at the University of Washington are co-inventors on the provisional patent application MBHB 23-1545-US-PRO (submitted) that incorporates the discoveries of IL-6R/IL1R/GP130 minibinders described in this manuscript. The remaining authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
- R01 HL168255/HL/NHLBI NIH HHS/United States
- R01 AG063845/AG/NIA NIH HHS/United States
- C19 HHMI INITIATIVE/Howard Hughes Medical Institute (HHMI)
- P30 GM133894/GM/NIGMS NIH HHS/United States
- R21 HL167211/HL/NHLBI NIH HHS/United States
- U01 HL169361/HL/NHLBI NIH HHS/United States
- R01 CA114536/CA/NCI NIH HHS/United States
- F30 HL160055/HL/NHLBI NIH HHS/United States
- R01 AI051321/AI/NIAID NIH HHS/United States
- R01 HL133308/HL/NHLBI NIH HHS/United States
- HR0011-21-2-0012/United States Department of Defense | Defense Advanced Research Projects Agency (DARPA)
- R01 CA240339/CA/NCI NIH HHS/United States
- HR0011835403 contract FA8750-17-C-0219/United States Department of Defense | Defense Advanced Research Projects Agency (DARPA)
- HDTRA1-21-1-0038/United States Department of Defense | Defense Threat Reduction Agency (DTRA)
LinkOut - more resources
Full Text Sources
Research Materials

