SpatialOne: end-to-end analysis of visium data at scale
- PMID: 39152991
- PMCID: PMC11374018
- DOI: 10.1093/bioinformatics/btae509
SpatialOne: end-to-end analysis of visium data at scale
Abstract
Motivation: Spatial transcriptomics allow to quantify mRNA expression within the spatial context. Nonetheless, in-depth analysis of spatial transcriptomics data remains challenging and difficult to scale due to the number of methods and libraries required for that purpose.
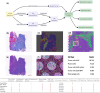
Results: Here we present SpatialOne, an end-to-end pipeline designed to simplify the analysis of 10x Visium data by combining multiple state-of-the-art computational methods to segment, deconvolve, and quantify spatial information; this approach streamlines the analysis of reproducible spatial-data at scale.
Availability and implementation: SpatialOne source code and execution examples are available at https://github.com/Sanofi-Public/spatialone-pipeline, experimental data is available at https://zenodo.org/records/12605154. SpatialOne is distributed as a docker container image.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
All authors are Sanofi employees and may hold shares and/or stock options in the company. The SpatialOne team is not connected in any way to 10x Genomics.
Figures
References
-
- Graham S, Vu QD, Raza SEA. et al. Hover-net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal 2019;58:101563. - PubMed
-
- Kasemeier-Kulesa JC, Morrison JA, McKinney S. et al. Cell-type profiling of the sympathetic nervous system using spatial transcriptomics and spatial mapping of mrna. Dev Dyn 2023;252:1130–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

