Exploring antidiabetic drug targets as potential disease-modifying agents in osteoarthritis
- PMID: 39153411
- PMCID: PMC11378937
- DOI: 10.1016/j.ebiom.2024.105285
Exploring antidiabetic drug targets as potential disease-modifying agents in osteoarthritis
Abstract
Background: Osteoarthritis is a leading cause of disability, and disease-modifying osteoarthritis drugs (DMOADs) could represent a pivotal advancement in treatment. Identifying the potential of antidiabetic medications as DMOADs could impact patient care significantly.
Methods: We designed a comprehensive analysis pipeline involving two-sample Mendelian Randomization (MR) (genetic proxies for antidiabetic drug targets), summary-based MR (SMR) (for mRNA), and colocalisation (for drug-target genes) to assess their causal relationship with 12 osteoarthritis phenotypes. Summary statistics from the largest genome-wide association meta-analysis (GWAS) of osteoarthritis and gene expression data from the eQTLGen consortium were utilised.
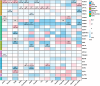
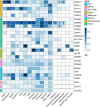
Findings: Seven out of eight major types of clinical antidiabetic medications were identified, resulting in fourteen potential drug targets. Sulfonylurea targets ABCC8/KCNJ11 were associated with increased osteoarthritis risk at any site (odds ratio (OR): 2.07, 95% confidence interval (CI): 1.50-2.84, P < 3 × 10-4), while PPARG, influenced by thiazolidinediones (TZDs), was associated with decreased risk of hand (OR: 0.61, 95% CI: 0.48-0.76, P < 3 × 10-4), finger (OR: 0.50, 95% CI: 0.35-0.73, P < 3 × 10-4), and thumb (OR: 0.49, 95% CI: 0.34-0.71, P < 3 × 10-4) osteoarthritis. Metformin and GLP1-RA, targeting GPD1 and GLP1R respectively, were associated with reduced risk of knee and finger osteoarthritis. In the SMR analyses, gene expression of KCNJ11, GANAB, ABCA1, and GSTP1, targeted by antidiabetic drugs, was significantly linked to at least one osteoarthritis phenotype and was replicated across at least two gene expression datasets. Additionally, increased KCNJ11 expression was related to decreased osteoarthritis risk and co-localised with at least one osteoarthritis phenotype.
Interpretation: Our findings suggest a potential therapeutic role for antidiabetic drugs in treating osteoarthritis. The results indicate that certain antidiabetic drug targets may modify disease progression, with implications for developing targeted DMOADs.
Funding: This study was funded by the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant (2022), the Shanghai Municipal Health Commission Health Industry Clinical Research Project (Grant No. 20224Y0139), Beijing Natural Science Foundation (Grant No. 7244458), and the Postdoctoral Fellowship Program (Grade C) of China Postdoctoral Science Foundation (Grant No. GZC20230130).
Keywords: Antidiabetic drugs; Drug targets; Gene expression; Mendelian randomization; Osteoarthritis.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Professor David J. Hunter provides consulting advice to Merck Serono, Pfizer, Lilly, TLCBio, and Novartis, outside the submitted work.
Figures



References
-
- Hunter D.J., Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393(10182):1745–1759. - PubMed
-
- Mobasheri A., Batt M. An update on the pathophysiology of osteoarthritis. Ann Phys Rehabil Med. 2016;59(5-6):333–339. - PubMed
-
- Mobasheri A., Rayman M.P., Gualillo O., Sellam J., van der Kraan P., Fearon U. The role of metabolism in the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2017;13(5):302–311. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

