Discovery of an embryonically derived bipotent population of endothelial-macrophage progenitor cells in postnatal aorta
- PMID: 39154007
- PMCID: PMC11330468
- DOI: 10.1038/s41467-024-51637-7
Discovery of an embryonically derived bipotent population of endothelial-macrophage progenitor cells in postnatal aorta
Abstract
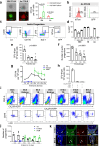
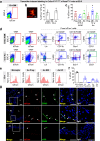
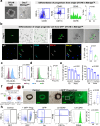
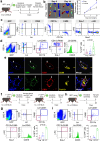
Converging evidence indicates that extra-embryonic yolk sac is the source of both macrophages and endothelial cells in adult mouse tissues. Prevailing views are that these embryonically derived cells are maintained after birth by proliferative self-renewal in their differentiated states. Here we identify clonogenic endothelial-macrophage (EndoMac) progenitor cells in the adventitia of embryonic and postnatal mouse aorta, that are independent of Flt3-mediated bone marrow hematopoiesis and derive from an early embryonic CX3CR1+ and CSF1R+ source. These bipotent progenitors are proliferative and vasculogenic, contributing to adventitial neovascularization and formation of perfused blood vessels after transfer into ischemic tissue. We establish a regulatory role for angiotensin II, which enhances their clonogenic and differentiation properties and rapidly stimulates their proliferative expansion in vivo. Our findings demonstrate that embryonically derived EndoMac progenitors participate in local vasculogenic responses in the aortic wall by contributing to the expansion of endothelial cells and macrophages postnatally.
© 2024. The Author(s).
Conflict of interest statement
S.J.N. has received research support from AstraZeneca, Amgen, Anthera, Eli Lilly, Esperion, Novartis, Cerenis, The Medicines Company, Resverlogix, InfraReDx, Roche, Sanofi-Regeneron and Liposcience and is a consultant for AstraZeneca, Akcea, Eli Lilly, Anthera, Kowa, Omthera, Merck, Takeda, Resverlogix, Sanofi-Regeneron, CSL Behring, Esperion and Boehringer Ingelheim. P.J.P. has received research support from Abbott Vascular, consulting fees from Amgen, Esperion, Eli Lilly, Novartis, Novo Nordisk and Sanofi, and speaker honoraria from Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Merck Schering-Plough, Pfizer, Novartis, Novo Nordisk and Sanofi. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- CDF 1161506/Department of Health | National Health and Medical Research Council (NHMRC)
- PG 1086796/Department of Health | National Health and Medical Research Council (NHMRC)
- IG 2001541/Department of Health | National Health and Medical Research Council (NHMRC)
- PRF 1111630/Department of Health | National Health and Medical Research Council (NHMRC)
- 100412/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

