Jumonji histone demethylases are therapeutic targets in small cell lung cancer
- PMID: 39154123
- PMCID: PMC11405284
- DOI: 10.1038/s41388-024-03125-x
Jumonji histone demethylases are therapeutic targets in small cell lung cancer
Abstract
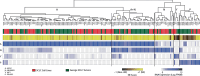
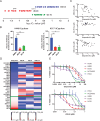
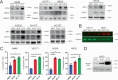
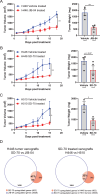
Small cell lung cancer (SCLC) is a recalcitrant cancer of neuroendocrine (NE) origin. Changes in therapeutic approaches against SCLC have been lacking over the decades. Here, we use preclinical models to identify a new therapeutic vulnerability in SCLC consisting of the targetable Jumonji lysine demethylase (KDM) family. We show that Jumonji demethylase inhibitors block malignant growth and that etoposide-resistant SCLC cell lines are particularly sensitive to Jumonji inhibition. Mechanistically, small molecule-mediated inhibition of Jumonji KDMs activates endoplasmic reticulum (ER) stress genes, upregulates ER stress signaling, and triggers apoptotic cell death. Furthermore, Jumonji inhibitors decrease protein levels of SCLC NE markers INSM1 and Secretogranin-3 and of driver transcription factors ASCL1 and NEUROD1. Genetic knockdown of KDM4A, a Jumonji demethylase highly expressed in SCLC and a known regulator of ER stress genes, induces ER stress response genes, decreases INSM1, Secretogranin-3, and NEUROD1 and inhibits proliferation of SCLC in vitro and in vivo. Lastly, we demonstrate that two different small molecule Jumonji KDM inhibitors (pan-inhibitor JIB-04 and KDM4 inhibitor SD70) block the growth of SCLC tumor xenografts in vivo. Our study highlights the translational potential of Jumonji KDM inhibitors against SCLC, a clinically feasible approach in light of recently opened clinical trials evaluating this drug class, and establishes KDM4A as a relevant target across SCLC subtypes.
© 2024. The Author(s).
Conflict of interest statement
JDM receives royalties from the NIH and UTSW for the distribution of human tumor cell lines. None of the other authors have any competing interests to declare.
Figures


References
-
- Brahmer JR, Lee JS, Ciuleanu TE, Bernabe Caro R, Nishio M, Urban L, et al. Five-year survival outcomes with nivolumab plus ipilimumab versus chemotherapy as first-line treatment for metastatic non-small-cell lung cancer in CheckMate 227. J Clin Oncol. 2023;41:1200–12. 10.1200/JCO.22.01503 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

