The Chlamydia effector IncE employs two short linear motifs to reprogram host vesicle trafficking
- PMID: 39154341
- PMCID: PMC12108946
- DOI: 10.1016/j.celrep.2024.114624
The Chlamydia effector IncE employs two short linear motifs to reprogram host vesicle trafficking
Abstract
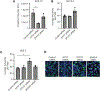
Chlamydia trachomatis, a leading cause of bacterial sexually transmitted infections, creates a specialized intracellular replicative niche by translocation and insertion of a diverse array of effectors (Incs [inclusion membrane proteins]) into the inclusion membrane. Here, we characterize IncE, a multifunctional Inc that encodes two non-overlapping short linear motifs (SLiMs) within its short cytosolic C terminus. The proximal SLiM, by mimicking just a small portion of an R-N-ethylmaleimide-sensitive factor adaptor protein receptor (SNARE) motif, binds and recruits syntaxin (STX)7- and STX12-containing vesicles to the inclusion. The distal SLiM mimics the sorting nexin (SNX)5 and SNX6 cargo binding site to recruit SNX6-containing vesicles to the inclusion. By simultaneously binding two distinct vesicle classes, IncE brings these vesicles in close apposition with each other at the inclusion to facilitate C. trachomatis intracellular development. Our work suggests that Incs may have evolved SLiMs to enable rapid evolution in a limited protein space to disrupt host cell processes.
Keywords: CP: Microbiology; Chlamydia trachomatis; SNARE; SNARE protein; host-pathogen interactions; inclusion membrane protein; intracellular bacteria; microbial pathogenesis; short linear motif; sorting nexin; syntaxin; vesicular trafficking.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


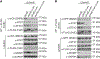



Update of
-
The Chlamydia effector IncE employs two short linear motifs to reprogram host vesicle trafficking.bioRxiv [Preprint]. 2024 Apr 24:2024.04.23.590830. doi: 10.1101/2024.04.23.590830. bioRxiv. 2024. Update in: Cell Rep. 2024 Aug 27;43(8):114624. doi: 10.1016/j.celrep.2024.114624. PMID: 38712241 Free PMC article. Updated. Preprint.
References
-
- Rockey DD, Scidmore MA, Bannantine JP, and Brown WJ (2002). Proteins in the chlamydial inclusion membrane. Microbes Infect. 4, 333–340. - PubMed
-
- Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, Von Dollen J, Rosenberg O, Gulbahce N, Jang G, Johnson T, et al. (2015). Global Mapping of the Inc-Human Interactome Reveals that Retromer Restricts Chlamydia Infection. Cell Host Microbe 18, 109–121. 10.1016/j.chom.2015.06.004. - DOI - PMC - PubMed
-
- Simonetti B, Paul B, Chaudhari K, Weeratunga S, Steinberg F, Gorla M, Heesom KJ, Bashaw GJ, Collins BM, and Cullen PJ (2019). Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins. Nat. Cell Biol 21, 1219–1233. 10.1038/s41556-019-0393-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

