NMR metabolomics-guided DNA methylation mortality predictors
- PMID: 39154540
- PMCID: PMC11378104
- DOI: 10.1016/j.ebiom.2024.105279
NMR metabolomics-guided DNA methylation mortality predictors
Abstract
Background: 1H-NMR metabolomics and DNA methylation in blood are widely known biomarkers predicting age-related physiological decline and mortality yet exert mutually independent mortality and frailty signals.
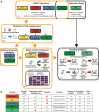
Methods: Leveraging multi-omics data in four Dutch population studies (N = 5238, ∼40% of which male) we investigated whether the mortality signal captured by 1H-NMR metabolomics could guide the construction of DNA methylation-based mortality predictors.
Findings: We trained DNA methylation-based surrogates for 64 metabolomic analytes and found that analytes marking inflammation, fluid balance, or HDL/VLDL metabolism could be accurately reconstructed using DNA-methylation assays. Interestingly, a previously reported multi-analyte score indicating mortality risk (MetaboHealth) could also be accurately reconstructed. Sixteen of our derived surrogates, including the MetaboHealth surrogate, showed significant associations with mortality, independent of relevant covariates.
Interpretation: The addition of our metabolic analyte-derived surrogates to the well-established epigenetic clock GrimAge demonstrates that our surrogates potentially represent valuable mortality signal.
Funding: BBMRI-NL, X-omics, VOILA, Medical Delta, NWO, ERC.
Keywords: Ageing biomarkers; DNA methylation predictors; Epidemiology; Epigenetic clock; Metabolic risk score; NMR metabolomics.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Authors declare no competing interests.
Figures





References
-
- Partridge L., Deelen J., Slagboom P.E. Facing up to the global challenges of ageing. Nature. 2018;561:45–56. - PubMed
-
- Comfort A. Test-Battery to measure ageing-rate in man. Lancet. 1969;294:1411–1415. - PubMed
-
- Blackburn E.H., Greider C.W., Szostak J.W. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

