CDK12 controls transcription at damaged genes and prevents MYC-induced transcription-replication conflicts
- PMID: 39155303
- PMCID: PMC11330984
- DOI: 10.1038/s41467-024-51229-5
CDK12 controls transcription at damaged genes and prevents MYC-induced transcription-replication conflicts
Abstract
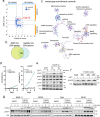
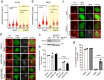
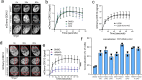
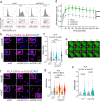
The identification of genes involved in replicative stress is key to understanding cancer evolution and to identify therapeutic targets. Here, we show that CDK12 prevents transcription-replication conflicts (TRCs) and the activation of cytotoxic replicative stress upon deregulation of the MYC oncogene. CDK12 was recruited at damaged genes by PARP-dependent DDR-signaling and elongation-competent RNAPII, to repress transcription. Either loss or chemical inhibition of CDK12 led to DDR-resistant transcription of damaged genes. Loss of CDK12 exacerbated TRCs in MYC-overexpressing cells and led to the accumulation of double-strand DNA breaks, occurring between co-directional early-replicating regions and transcribed genes. Overall, our data demonstrate that CDK12 protects genome integrity by repressing transcription of damaged genes, which is required for proper resolution of DSBs at oncogene-induced TRCs. This provides a rationale that explains both how CDK12 deficiency can promote tandem duplications of early-replicated regions during tumor evolution, and how CDK12 targeting can exacerbate replicative-stress in tumors.
© 2024. The Author(s).
Conflict of interest statement
S.C. and L.C. declare a patent application (IT102021000017414, PCT/IB2022/056112, grant date 03/08/2023). The remaining Authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

