PRRSV hijacks DDX3X protein and induces ferroptosis to facilitate viral replication
- PMID: 39155369
- PMCID: PMC11331664
- DOI: 10.1186/s13567-024-01358-y
PRRSV hijacks DDX3X protein and induces ferroptosis to facilitate viral replication
Abstract
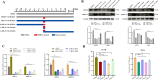
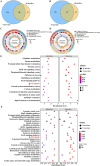
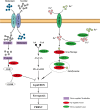
Porcine reproductive and respiratory syndrome virus (PRRSV) is a severe disease with substantial economic consequences for the swine industry. The DEAD-box helicase 3 (DDX3X) is an RNA helicase that plays a crucial role in regulating RNA metabolism, immunological response, and even RNA virus infection. However, it is unclear whether it contributes to PRRSV infection. Recent studies have found that the expression of DDX3X considerably increases in Marc-145 cells when infected with live PRRSV strains Ch-1R and SD16; however, it was observed that inactivated viruses did not lead to any changes. By using the RK-33 inhibitor or DDX3X-specific siRNAs to reduce DDX3X expression, there was a significant decrease in the production of PRRSV progenies. In contrast, the overexpression of DDX3X in host cells substantially increased the proliferation of PRRSV. A combination of transcriptomics and metabolomics investigations revealed that in PRRSV-infected cells, DDX3X gene silencing severely affected biological processes such as ferroptosis, the FoxO signalling pathway, and glutathione metabolism. The subsequent transmission electron microscopy (TEM) imaging displayed the typical ferroptosis features in PRRSV-infected cells, such as mitochondrial shrinkage, reduction or disappearance of mitochondrial cristae, and cytoplasmic membrane rupture. Conversely, the mitochondrial morphology was unchanged in DDX3X-inhibited cells. Furthermore, silencing of the DDX3X gene changed the expression of ferroptosis-related genes and inhibited the virus proliferation, while the drug-induced ferroptosis inversely promoted PRRSV replication. In summary, these results present an updated perspective of how PRRSV infection uses DDX3X for self-replication, potentially leading to ferroptosis via various mechanisms that promote PRRSV replication.
Keywords: DDX3X; PRRSV; ferroptosis; metabolome; transcriptome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







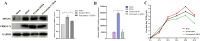
References
MeSH terms
Substances
Grants and funding
- K4030218170/Xi'an IRIS Livestock Technology Co. Ltd.
- 2017M610659/China Postdoctoral Science Foundation
- 2018T111113/China Postdoctoral Science Foundation
- SKLVEB2016KFKT014/State Key Laboratory of Drug Research, Chinese Academy of Sciences
- 2452019053/Fundamental Research Funds for the Central Universities
LinkOut - more resources
Full Text Sources

