Hypoxia-inducible factor-1α promotes macrophage functional activities in protecting hypoxia-tolerant large yellow croaker (Larimichthys crocea) against Aeromonas hydrophila infection
- PMID: 39156889
- PMCID: PMC11327042
- DOI: 10.3389/fimmu.2024.1410082
Hypoxia-inducible factor-1α promotes macrophage functional activities in protecting hypoxia-tolerant large yellow croaker (Larimichthys crocea) against Aeromonas hydrophila infection
Abstract
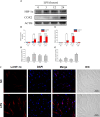
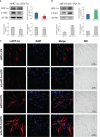
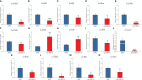
The immune system requires a high energy expenditure to resist pathogen invasion. Macrophages undergo metabolic reprogramming to meet these energy requirements and immunologic activity and polarize to M1-type macrophages. Understanding the metabolic pathway switching in large yellow croaker (Larimichthys crocea) macrophages in response to lipopolysaccharide (LPS) stimulation and whether this switching affects immunity is helpful in explaining the stronger immunity of hypoxia-tolerant L. crocea. In this study, transcript levels of glycolytic pathway genes (Glut1 and Pdk1), mRNA levels or enzyme activities of glycolytic enzymes [hexokinase (HK), phosphofructokinase (PFK), pyruvate kinase (PK), and lactate dehydrogenase A (LDHA)], aerobic respiratory enzymes [pyruvate dehydrogenase (PDH), isocitrate dehydrogenase (IDH), and succinate dehydrogenase (SDH)], metabolites [lactic acid (LA) and adenosine triphosphate (ATP)], levels of bactericidal products [reactive oxygen species (ROS) and nitric oxide (NO)], and transcripts and level changes of inflammatory factors [IL1β, TNFα, and interferon (IFN) γ] were detected in LPS-stimulated L. crocea head kidney macrophages. We showed that glycolysis was significantly induced, the tricarboxylic acid (TCA) cycle was inhibited, and metabolic reprogramming occurred, showing the Warburg effect when immune cells were activated. To determine the potential regulatory mechanism behind these changes, LcHIF-1α was detected and found to be significantly induced and transferred to the nucleus after LPS stimulation. LcHif-1α interference led to a significant reduction in glycolytic pathway gene transcript expression, enzyme activity, metabolites, bactericidal substances, and inflammatory factor levels; a significant increase in the aerobic respiration enzymes; and decreased migration, invasion, and phagocytosis. Further ultrastructural observation by electron microscopy showed that fewer microspheres contained phagocytes and that more cells were damaged after LcHif-1α interference. LcHif-1α overexpression L. crocea head kidney macrophages showed the opposite trend, and promoter activities of Ldha and Il1β were significantly enhanced after LcHif-1α overexpression in HEK293T cells. Our data showed that LcHIF-1α acted as a metabolic switch in L. crocea macrophages and was important in polarization. Hypoxia-tolerant L. crocea head kidney showed a stronger Warburg effect and inhibited the TCA cycle, higher metabolites, and bactericidal substance levels. These results collectively revealed that LcHif-1α may promote the functional activities of head kidney macrophages in protecting hypoxia-tolerant L. crocea from Aeromonas hydrophila infection.
Keywords: Larimichthys crocea; glycolysis; immunity; macrophages; metabolic reprogramming.
Copyright © 2024 Zhang, Wang, Gao, Li, Meng, Wu, Ding, Shen and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Zhang ZX, Wang ZY, Fang M, Ye K, Tang X, Zhang DL. Genome-wide association analysis on host resistance against the rotten body disease in a naturally infected population of large yellow croaker larimichthys crocea . Aquaculture. (2022) 548:737615. doi: 10.1016/j.aquaculture.2021.737615 - DOI
-
- Ding J, Zhang Y, Wang J, Liu C, Gao X, Wu Y, et al. . Genome-wide association study identified candidate snps and genes associated with hypoxia tolerance in large yellow croaker (Larimichthys crocea). Aquaculture. (2022) 560:738472. doi: 10.1016/j.aquaculture.2022.738472 - DOI
-
- Ding J, Liu C, Luo S, Zhang Y, Gao X, Wu X, et al. . Transcriptome and physiology analysis identify key metabolic changes in the liver of the large yellow croaker (Larimichthys crocea) in response to acute hypoxia. Ecotoxicol Environ Saf. (2020) 189:109957. doi: 10.1016/j.ecoenv.2019.109957 - DOI - PubMed
-
- Ding J, Gao Z, Wang J, Zhang Y, Wang X, Wu X, et al. . Genome-wide association and transcriptome analysis provide the snps and molecular insights into the hypoxia tolerance in large yellow croaker (Larimichthys crocea). Aquaculture. (2023) 573:739547. doi: 10.1016/j.aquaculture.2023.739547 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

