Bat humoral immunity and its role in viral pathogenesis, transmission, and zoonosis
- PMID: 39156901
- PMCID: PMC11329927
- DOI: 10.3389/fimmu.2024.1269760
Bat humoral immunity and its role in viral pathogenesis, transmission, and zoonosis
Abstract
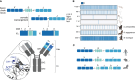
Bats harbor viruses that can cause severe disease and death in humans including filoviruses (e.g., Ebola virus), henipaviruses (e.g., Hendra virus), and coronaviruses (e.g., SARS-CoV). Bats often tolerate these viruses without noticeable adverse immunological effects or succumbing to disease. Previous studies have largely focused on the role of the bat's innate immune response to control viral pathogenesis, but little is known about bat adaptive immunity. A key component of adaptive immunity is the humoral response, comprised of antibodies that can specifically recognize viral antigens with high affinity. The antibody genes within the 1,400 known bat species are highly diverse, and these genetic differences help shape fundamental aspects of the antibody repertoire, including starting diversity and viral antigen recognition. Whether antibodies in bats protect, mediate viral clearance, and prevent transmission within bat populations is poorly defined. Furthermore, it is unclear how neutralizing activity and Fc-mediated effector functions contribute to bat immunity. Although bats have canonical Fc genes (e.g., mu, gamma, alpha, and epsilon), the copy number and sequences of their Fc genes differ from those of humans and mice. The function of bat antibodies targeting viral antigens has been speculated based on sequencing data and polyclonal sera, but functional and biochemical data of monoclonal antibodies are lacking. In this review, we summarize current knowledge of bat humoral immunity, including variation between species, their potential protective role(s) against viral transmission and replication, and address how these antibodies may contribute to population dynamics within bats communities. A deeper understanding of bat adaptive immunity will provide insight into immune control of transmission and replication for emerging viruses with the potential for zoonotic spillover.
Keywords: Chiroptera; bat immunity; humoral responses; immunoglobulin repertoire; infectious diseases.
Copyright © 2024 Roffler, Maurer, Lunn, Sironen, Forbes and Schmidt.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

