Progress in Research on Inhibitors Targeting SARS-CoV-2 Main Protease (Mpro)
- PMID: 39157135
- PMCID: PMC11325518
- DOI: 10.1021/acsomega.4c03023
Progress in Research on Inhibitors Targeting SARS-CoV-2 Main Protease (Mpro)
Abstract
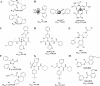
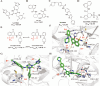
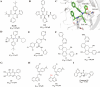
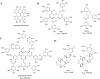
Since 2019, the novel coronavirus (SARS-CoV-2) has caused significant morbidity and millions of deaths worldwide. The Coronavirus Disease 2019 (COVID-19), caused by SARS-CoV-2 and its variants, has further highlighted the urgent need for the development of effective therapeutic agents. Currently, the highly conserved and broad-spectrum nature of main proteases (Mpro) renders them of great importance in the field of inhibitor study. In this study, we categorize inhibitors targeting Mpro into three major groups: mimetic, nonmimetic, and natural inhibitors. We then present the research progress of these inhibitors in detail, including their mechanism of action, antiviral activity, pharmacokinetic properties, animal experiments, and clinical studies. This review aims to provide valuable insights and potential avenues for the development of more effective antiviral drugs against SARS-CoV-2.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.J Virol. 2022 Apr 27;96(8):e0201321. doi: 10.1128/jvi.02013-21. Epub 2022 Apr 7. J Virol. 2022. PMID: 35389231 Free PMC article.
-
Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.J Virol. 2022 Jan 12;96(1):e0125321. doi: 10.1128/JVI.01253-21. Epub 2021 Sep 29. J Virol. 2022. PMID: 34586857 Free PMC article.
-
Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy.ACS Comb Sci. 2020 Jun 8;22(6):297-305. doi: 10.1021/acscombsci.0c00058. Epub 2020 May 27. ACS Comb Sci. 2020. PMID: 32402186 Review.
-
Adaptive Mutation in the Main Protease Cleavage Site of Feline Coronavirus Renders the Virus More Resistant to Main Protease Inhibitors.J Virol. 2022 Sep 14;96(17):e0090722. doi: 10.1128/jvi.00907-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000844 Free PMC article.
-
Progress of SARS-CoV-2 Main protease peptide-like inhibitors.Chem Biol Drug Des. 2024 Jan;103(1):e14425. doi: 10.1111/cbdd.14425. Epub 2023 Dec 11. Chem Biol Drug Des. 2024. PMID: 38082476 Review.
Cited by
-
A Reflection on the Use of Molecular Simulation to Respond to SARS-CoV-2 Pandemic Threats.J Phys Chem Lett. 2025 Apr 3;16(13):3249-3263. doi: 10.1021/acs.jpclett.4c03654. Epub 2025 Mar 21. J Phys Chem Lett. 2025. PMID: 40118074 Free PMC article. Review.
-
Identification of potential COVID-19 Mpro inhibitors through covalent drug docking, molecular dynamics simulation, and MMGBSA calculation.Sci Rep. 2025 Jul 1;15(1):20500. doi: 10.1038/s41598-025-05375-5. Sci Rep. 2025. PMID: 40596055 Free PMC article.
-
Research Progress on the Structure and Function, Immune Escape Mechanism, Antiviral Drug Development Methods, and Clinical Use of SARS-CoV-2 Mpro.Molecules. 2025 Jan 16;30(2):351. doi: 10.3390/molecules30020351. Molecules. 2025. PMID: 39860219 Free PMC article. Review.
References
-
- Zou L.; Ruan F.; Huang M.; Liang L.; Huang H.; Hong Z.; Yu J.; Kang M.; Song Y.; Xia J.; Guo Q.; Song T.; He J.; Yen H. L.; Peiris M.; Wu J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J. Med. 2020, 382 (12), 1177–1179. 10.1056/NEJMc2001737. - DOI - PMC - PubMed
-
- Lu N.; Gu T.; Tian X.; Zhao S.; Jin G.; Mangaladoss F.; Qiao Y.; Liu K.; Zhao R.; Dong Z. Acetylshikonin inhibits inflammatory responses and Papain-like protease activity in murine model of COVID-19. Signal transduction and targeted therapy 2022, 7 (1), 371.10.1038/s41392-022-01220-7. - DOI - PMC - PubMed
-
- Wang Q.; Iketani S.; Li Z.; Liu L.; Guo Y.; Huang Y.; Bowen A. D.; Liu M.; Wang M.; Yu J.; Valdez R.; Lauring A. S.; Sheng Z.; Wang H. H.; Gordon A.; Liu L.; Ho D. D. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2023, 186 (2), 279–286. 10.1016/j.cell.2022.12.018. - DOI - PMC - PubMed
-
- Carabelli A. M.; Peacock T. P.; Thorne L. G.; Harvey W. T.; Hughes J.; de Silva T. I.; Peacock S. J.; Barclay W. S.; de Silva T. I.; Towers G. J.; Robertson D. L. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol 2023, 21 (3), 162–177. 10.1038/s41579-022-00841-7. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
