Based on mutated aptamer-smartphone colorimetric detection of metronidazole in milk
- PMID: 39157440
- PMCID: PMC11327025
- DOI: 10.3389/fbioe.2024.1444846
Based on mutated aptamer-smartphone colorimetric detection of metronidazole in milk
Abstract
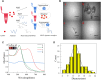
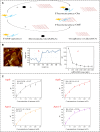
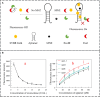
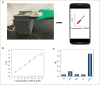
Excessive residue of metronidazole (MNZ) in food is harmful to the human body. There is an urgent demand to develop a portable tool for MNZ detection on-site. In this study, fifteen aptamers were prepared through targeted base mutation. Apt1-3 with the highest enrichment was chosen for further study. Its affinity was characterized by molecular docking simulation, AuNPs colorimetric assay, graphene oxide (GO) fluorescence assay, and exonuclease assay. Kd was determined by GO fluorescence assay (Kd: 92.60 ± 25.59 nM). Its specificity was also characterized by an exonuclease assay. A novel aptasensor was constructed by using the newly identified aptamer combined with the smartphone dark box. The principle of color change is caused by the aggregation state of AuNPs. Smartphones act as reading instruments. The detection can be completed in just a few seconds without the aid of instruments, achieving a detection limit of 0.15 nmol/mL and a range of 6.7-44.4 nmol/mL (R 2 = 0.9810). Therefore, the constructed smartphone colorimetric sensor based on mutant aptamers has important applications in food detection.
Keywords: aptamer; base mutation; colorimetric; metronidazole; smartphone.
Copyright © 2024 Zhang, Qin, Yuan, Wang, Yao and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

