An unbiased lncRNA dropout CRISPR-Cas9 screen reveals RP11-350G8.5 as a novel therapeutic target for multiple myeloma
- PMID: 39158066
- PMCID: PMC11522894
- DOI: 10.1182/blood.2023021991
An unbiased lncRNA dropout CRISPR-Cas9 screen reveals RP11-350G8.5 as a novel therapeutic target for multiple myeloma
Abstract
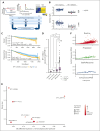
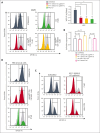
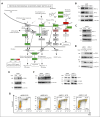
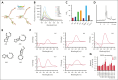
Multiple myeloma (MM) is an incurable malignancy characterized by altered expression of coding and noncoding genes promoting tumor growth and drug resistance. Although the crucial role of long noncoding RNAs (lncRNAs) in MM is clearly established, the function of the noncoding RNAome, which might allow the design of novel therapeutics, is largely unknown. We performed an unbiased CRISPR-Cas9 loss-of-function screen of 671 lncRNAs in MM cells and their bortezomib (BZB)-resistant derivative. To rank functionally and clinically relevant candidates, we designed and used a bioinformatic prioritization pipeline combining functional data from cellular screens with prognostic and transcriptional data from patients with MM. With this approach, we unveiled and prioritized 8 onco-lncRNAs essential for MM cell fitness, associated with high expression and poor prognosis in patients with MM. The previously uncharacterized RP11-350G8.5 emerged as the most promising target, irrespective of BZB resistance. We (1) demonstrated the anti-tumoral effect obtained by RP11-350G8.5 inhibition in vitro and in vivo; (2) highlighted a modulation of the unfolded protein response and the induction of immunogenic cell death triggered by the RP11-350G8.5 knockout, via RNA sequencing and molecular studies; (3) characterized its cytoplasmic homing through RNA fluorescence in situ hybridization; and (4) predicted its 2-dimensional structure and identified 2 G-quadruplex and 3 hairpin-forming regions by biophysical assays, including thioflavin T, 1H nuclear magnetic resonance, and circular dichroism, to pave the way to the development of novel targeted therapeutics. Overall, we provided innovative insights about unexplored lncRNAs in MM and identified RP11-350G8.5 as an oncogenic target for treatment-naïve and BZB-resistant patients with MM.
© 2024 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: F.I. receives funds from Open Targets, a public-private initiative involving academia and industry, and performs consultancy for the Cancer Research Horizons–AstraZeneca Functional Genomics Centre and Mosaic Therapeutics. The remaining authors declare no competing financial interests.
Figures






References
-
- van de Donk N, Pawlyn C, Yong KL. Multiple myeloma. Lancet. 2021;397(10272):410–427. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

