Polygenic polymorphism is associated with NKG2A repertoire and influences lymphocyte phenotype and function
- PMID: 39158076
- PMCID: PMC11568789
- DOI: 10.1182/bloodadvances.2024013508
Polygenic polymorphism is associated with NKG2A repertoire and influences lymphocyte phenotype and function
Abstract
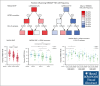
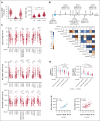
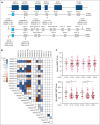
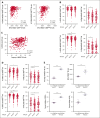
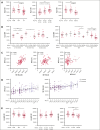
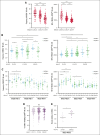
CD94/NKG2A is a heterodimeric receptor commonly found on natural killer (NK) and T cells, and its interaction with its ligand HLA-E on adjacent cells leads to inhibitory signaling and cell suppression. We have identified several killer cell lectin-like receptor (KLR)C1 (NKG2A) single nucleotide polymorphisms (SNPs) that are associated with NKG2A expression on NK cells, CD8+ T cells, and Vγ9/Vδ2+ T cells. Additionally, due to strong linkage disequilibrium, polymorphisms in KLRC2 (NKG2C) and KLRK1 (NKG2D) are also associated with NKG2A surface density and frequency. NKG2A surface expression correlates with single-cell NK responsiveness, and NKG2A+ NK cell frequency is associated with total NK repertoire response and inhibitability, making the identification of SNPs responsible for expression and frequency important for predicting the innate immune response. Because HLA-E expression is dependent on HLA class I signal peptides, we analyzed the relationship between peptide abundance and HLA-E expression levels. Our findings revealed a strong association between peptide availability and HLA-E expression. We identified the HLA-C killer immunoglobulin-like receptor ligand epitope as a predictive marker for HLA-ABC expression, with the HLA-C1 epitope associated with high HLA-E expression and the HLA-C2 epitope associated with low HLA-E expression. The relationship between HLA-C epitopes and HLA-E expression was independent of HLA-E allotypes and HLA-B leader peptides. Although HLA-E expression showed no significant influence on NKG2A-mediated NK education, it did affect NK cell inhibition. In summary, these findings underscore the importance of NKG2A SNPs and HLA-C epitopes as predictive markers of NK cell phenotype and function and should be evaluated as prognostic markers for diseases that express high levels of HLA-E.
© 2024 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: M.K.P. and K.C.H. are listed as inventors on a provisional patent for the design and use of HLA-E:peptide chimeric molecules (US provisional patent application number, 63/173,966). The remaining authors declare no competing financial interests.
Figures


References
-
- Angelini DF, Zambello R, Galandrini R, et al. NKG2A inhibits NKG2C effector functions of gammadelta T cells: implications in health and disease. J Leukoc Biol. 2011;89(1):75–84. - PubMed
-
- Raulet DH. Interplay of natural killer cells and their receptors with the adaptive immune response. Nat Immunol. 2004;5(10):996–1002. - PubMed
-
- Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436(7051):709–713. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

