Genetically predicted lipoprotein(a) associates with coronary artery plaque severity independent of low-density lipoprotein cholesterol
- PMID: 39158116
- PMCID: PMC12178393
- DOI: 10.1093/eurjpc/zwae271
Genetically predicted lipoprotein(a) associates with coronary artery plaque severity independent of low-density lipoprotein cholesterol
Abstract
Aims: Elevated lipoprotein(a) [Lp(a)] is a causal risk factor for atherosclerotic cardiovascular disease, but the mechanisms of risk are debated. Studies have found inconsistent associations between Lp(a) and measurements of atherosclerosis. We aimed to assess the relationship between Lp(a), low-density lipoprotein cholesterol (LDL-C), and coronary artery plaque severity.
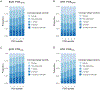
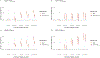
Methods and results: The study population consisted of participants of the Million Veteran Program who have undergone an invasive angiogram. The primary exposure was genetically predicted Lp(a) estimated by a polygenic score. Genetically predicted LDL-C was also assessed for comparison. The primary outcome was coronary artery plaque severity categorized as normal, non-obstructive disease, one-vessel disease, two-vessel disease, and three-vessel or left main disease. Among 18 927 adults of genetically inferred European ancestry and 4039 adults of genetically inferred African ancestry, we observed consistent associations between genetically predicted Lp(a) and obstructive coronary plaque, with effect sizes trending upward for increasingly severe categories of disease. Associations were independent of risk factors, clinically measured LDL-C and genetically predicted LDL-C. However, we did not find strong or consistent evidence for an association between genetically predicted Lp(a) and risk for non-obstructive plaque.
Conclusion: Genetically predicted Lp(a) is positively associated with coronary plaque severity independent of LDL-C, consistent with Lp(a) promoting atherogenesis. However, the effects of Lp(a) may be greater for progression of plaque to obstructive disease than for the initial development of non-obstructive plaque. A limitation of this study is that Lp(a) was estimated using genetic markers and could not be directly assayed nor could apo(a) isoform size.
Keywords: Atherosclerosis; Coronary artery disease; LDL-C; Lipoprotein(a); Polygenic score.
Plain language summary
This study assessed the association between genetic propensity towards higher lipoprotein(a) [Lp(a)] in the blood and the severity of coronary artery plaque seen on clinical angiograms, independent of other factors, including low-density lipoprotein cholesterol (LDL-C). The study was conducted in a large US population using data from the Million Veteran Program.Genetically predicted high Lp(a) was associated with obstructive coronary plaque, but it was not associated with non-obstructive coronary plaque. This association was independent of LDL-C, and the association was greater for more severe forms of disease.The mechanisms of association between Lp(a) and cardiovascular events are debated. Prior studies have shown that Lp(a) does not associate with early markers of atherosclerosis. Our analyses support the idea that Lp(a) plays less of a role in early plaque initiation, but plays a significant role in the progression of plaque towards more severe disease, independent of LDL-C.
© The Author(s) 2024. Published by Oxford University Press on behalf of the European Society of Cardiology.
Conflict of interest statement
Conflict of interest: J.A. Lynch reports research grants from the following for-profit organizations outside this submitted work: Alnylam Pharmaceuticals Inc., Astellas Pharma Inc., AstraZeneca Pharmaceuticals LP, Biodesix Inc., Celgene Corporation, Cerner Enviza, GSK PLC, IQVIA Inc., Janssen Pharmaceuticals Inc., Kantar Health, Myriad Genetic Laboratories Inc., Novartis International AG, and Parexel International Corporation through the University of Utah or Western Institute for Veteran Research.
Figures


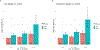
Comment in
-
The mysterious lipoprotein(a): moving towards further understanding of its atherogenic role.Eur J Prev Cardiol. 2025 Jan 27;32(2):128-130. doi: 10.1093/eurjpc/zwae321. Eur J Prev Cardiol. 2025. PMID: 39418090 No abstract available.
References
-
- Reyes-Soffer G, Ginsberg HN, Berglund L, et al. Lipoprotein(a): A Genetically Determined, Causal, and Prevalent Risk Factor for Atherosclerotic Cardiovascular Disease: A Scientific Statement From the American Heart Association. Arterioscler Thromb Vasc Biol. 2022;42(1):e48–e60. doi: 10.1161/ATV.0000000000000147 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

